Journal of Biology and Medicine
NGS analysis of unexplained Community-Acquired Pneumonia (CAP) cases in South Korea
Sooyeon Lim1-2*, Jae Kyung Lee4, Han Sol Lee1, Ji Yun Noh1-3, Joon Young Song1-3, Hee Jin Cheong1-3 and Woo Joo Kim1-3*
2Vaccine Innovation Center-KU Medicine, Seoul, Republic of Korea
3Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea
4Department of Biomedical Sciences, BK21 PLUS program, College of Medicine, Korea University Guro Hospital, Seoul 08308, Korea
Sooyeon Lim, PhD, Asia Pacific Influenza Institute, Korea University College of Medicine, Seoul, Republic of Korea, Guro Hospital, Gurodong-ro 148, Guro-gu, Seoul 08308, Republic of Korea. Tel.: +82 2 2626 1898; Fax: +82 2 2626 1105; E-mail: [email protected]
Cite this as
Lim S, Lee JK, Lee HS, Noh JY, Song JY, et al. (2023) NGS analysis of unexplained Community-Acquired Pneumonia (CAP) cases in South Korea. J Biol Med 7(1): 024-030. DOI: 10.17352/jbm.000038Copyright License
© 2023 Lim S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.In general, pneumonia has known to be closely associated with respiratory infection of viruses, bacteria, fungi, and parasites. In South Korea, pneumonia is a leading cause of death that continues to threaten public health every year. Through the tertiary hospital-based influenza surveillance system in South Korea, nasopharyngeal swab specimens were obtained from patients with unexplained cases of Community-Acquired Pneumonia (CAP) between 2011 and 2017. After real-time PCR screening using respiratory viral panels, the samples were found negative for 16 common respiratory pathogens including adenovirus, influenza viruses, rhinovirus, respiratory syncytial virus, coronavirus, metapneumovirus, and parainfluenza viruses. The aim of this study was to investigate the patient microbiota and examine the etiology of CAP requiring hospitalization. The nasopharyngeal microbiome of adult patients during CAP was analyzed using Next-Generation Sequencing (NGS) on the Illumina MiSeq platform and a subsequent bioinformatics pipeline. Viral nucleic acids were nearly absent in the samples and failed to generate any sequence reads. On the other hand, samples were enriched with a diverse bacterial community, which was mainly comprised of Corynebacterium, Staphylococcus, Streptococcus, Haemophilus, Moraxella, Acinetobacter, and Rhizobium genera. Despite the diversity of bacterial composition, only a few dominant species with > 1% abundance were identified in each patient sample. Population analysis at the genus level showed that microbial diversity varied according to age, sex, and location.
Introduction
Community-Acquired Pneumonia (CAP) is an infectious disease that affects the global population as one of the leading causes of death [1]. World Health Organization has ranked lower respiratory infections like pneumonia, estimated to have caused approximately 3 million deaths worldwide, as the third leading cause of death in 2015. In Korea, according to “Causes of Death Statistics” released by Statistics Korea (KOSTAT), pneumonia has remained one of the top ten leading causes of death in Korea since 1991, with a mortality rate that tends to increase annually [2]. The nasopharynx is a reservoir for various human pathogens responsible for respiratory infections, and the nasopharyngeal carriage of a specific arrangement of microorganisms has been used to determine an etiology for pneumonia [3]. As a result, understanding the nasopharyngeal microbiome is important in defining the main causative pathogens of respiratory infections like pneumonia. Unfortunately, not many studies have focused on investigating the nasopharyngeal carriage of pathogens during respiratory infections like pneumonia, in addition to investigating the microbial etiology of CAP in Korea. Therefore, as part of a tertiary hospital-based surveillance system of influenza, eight hospitals in Korea participated in a prospective cohort study from 2011 to 2017 [4]. Nasopharyngeal swabs were obtained from patients admitted to the hospital with CAP syndrome. Since clinical findings are often insufficient for etiological determination, microbiological investigations were performed to provide supporting information for appropriate diagnosis and treatment [5]. This study was conducted primarily to provide a comprehensive overview of the diversity present in the nasopharyngeal microbiota of patients with CAP utilizing the Next-Generation Sequencing (NGS) technique.
Methods and materials
Nasopharyngeal swabs, stored in VTM, were collected from CAP patients requiring hospitalization at one of the eight participating hospitals in the cohort study from 2011 to 2017. Specimens from the following hospitals were included in this study: Korea University Guro and Ansan Hospitals, Inha University Hospital, Chungbuk University Hospital, Wonju Severance Christian Hospital, Busan University Hospital, Kyungbuk University Hospital, and the Catholic University of Korea St.Vincent’s Hospital. The real-time PCR assay was performed as part of routine diagnostic testing for respiratory viruses. When negative results were confirmed for 16 common respiratory viruses, a total of 92 patient samples were selected for further analysis. The study was divided into two parts: real-time PCR analysis and NGS analysis utilizing the Illumina MiSeq platform.
Real-time PCR analysis
A real-time PCR technique using SYBR green was employed for targeted viral analysis. Primers were designed for the following viruses of interest: enterovirus D68, WU polyomavirus, KI polyomavirus, Pteropine orthoreovirus, and human parechovirus 1, 3, and 6. Primer sets used for qRT-PCR analysis are listed in Table 1 [6-10]. Negative results were confirmed for all viral targets from the Ct values and melting plots obtained for the samples. A total number of 92 samples were selected to proceed to the microbiome analysis step.
Virome analysis
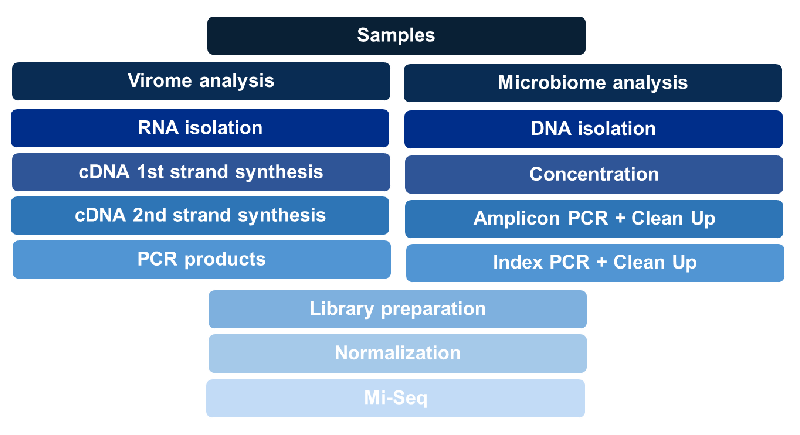
Samples were prepared using 0.45-µm-pore-size disc filters (Minisart Syringe Filter, Sartorius) for viral enrichment [11]. RNA isolation was performed using QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s protocol. Following first strand cDNA synthesis using V-N primer: 5’- GTT TCC CAG TCA CGA TC NNNNNN-3’. [12], second-strand cDNA was synthesized using Klenow Large Fragment (Takara). The resulting double-stranded DNA was then amplified in a 28-cycle PCR reaction. Amplicons were gel electrophoresed and analyzed by Bioanalyzer (Agilent) to confirm the presence of viral nucleic acid prior to next-generation sequencing (Figure 1).
Microbiome analysis
Bacterial 16s rRNA and viral screenings were carried out in the following manner: RNA/DNA isolation, PCR amplification, library preparation, and MiSeq sequencing and data analysis (Figure 1). Hypervariable regions V3 and V4 were selected for amplicon sequencing of the 16s rRNA gene [13,14]. DNA was extracted using a Power Soil DNA Isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) and concentrated using Hyper-Vac. The resulting nucleic acid was amplified using limited cycle PCR with Illumina-provided primers targeting the V3 and V4 region (Forward 5’-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3’; Reverse 5’-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C- 3’). V3 and V4 sequence reads were obtained using Nextera XT DNA library preparation kit and MiSeq Reagent Kit v3(Illumina). All quantifications of amplifications were performed using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc.), Agilent High Sensitivity DNA kit (Agilent Technologies, Inc.), and Xpose Compact Spectrometer (Trinean). Amplification products were purified using the Agencourt AMPure XP system (Beckman Coulter Inc.). Negative controls were included throughout the progression of this study. Amplicon libraries for both viral and bacterial analysis used 8% PhiX control (Illumina, Inc.).
Metagenomic analysis
The metagenome profile included the counts of reads that aligned to each contig, which could be analyzed using metagenomic tools and the R package (R Foundation for Statistical Computing, Vienna, Austria). The samples were then filtered based on the relative abundance of each OTU identified. The relative abundance was calculated based on the MiSeq sequence read data. Bacterial community profiles of the remaining samples included OTUs with >1% relative abundance. The Metagenomics workflow classifies bacteria from a metagenomic sample by amplifying specific regions in 16S ribosomal RNA. Reads are classified using a Greengenes taxonomy database of 16S ribosomal RNA data. The Metagenomics workflow demultiplexes indexed reads, generates FASTQ files, and then classifies reads and generated a classification of reads at several taxonomic levels: kingdom, phylum, class, order, family, and genus or species. The classification step uses ClassifyReads, a proprietary algorithm that provides a species-level classification for paired-end reads. This process involves matching short subsequences of the reads to a set of 16S reference sequences. The accumulated word matches for each read are used to assign reads to a particular taxonomic classification. Analysis results list the total number of classified clusters for each sample at each taxonomic level.
Results
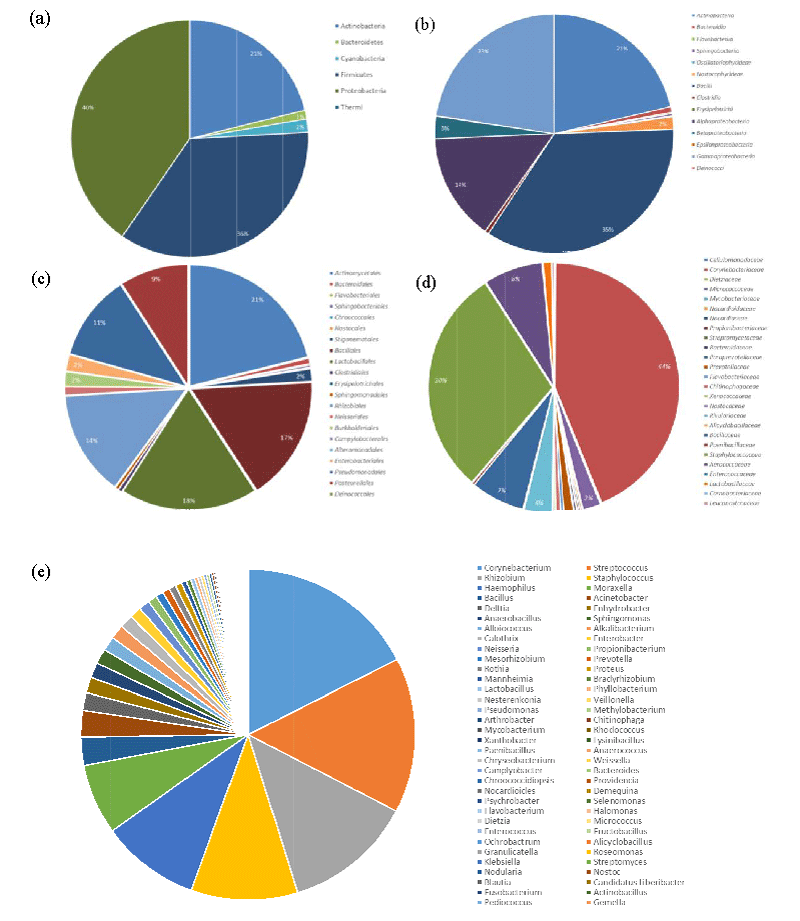
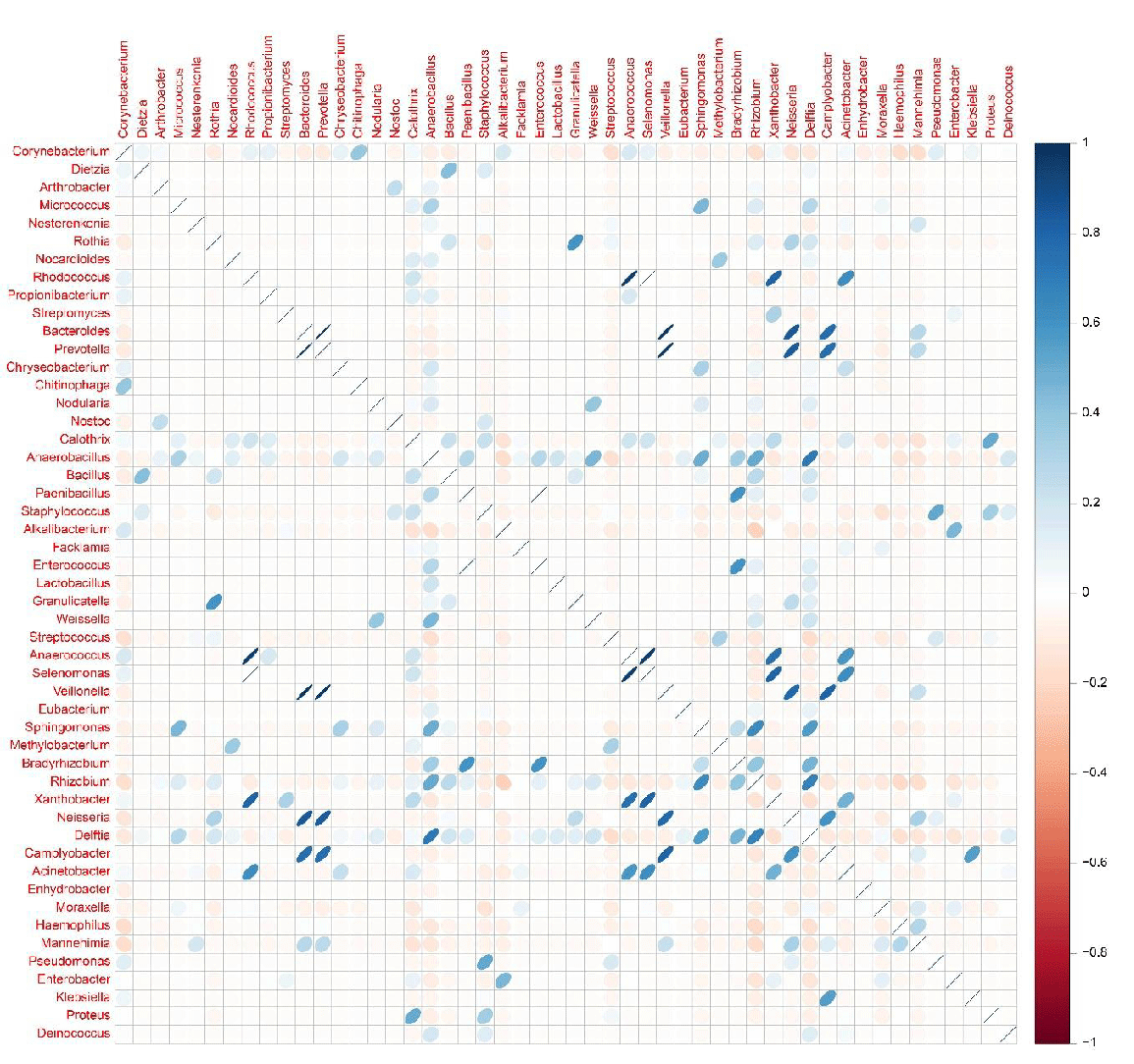
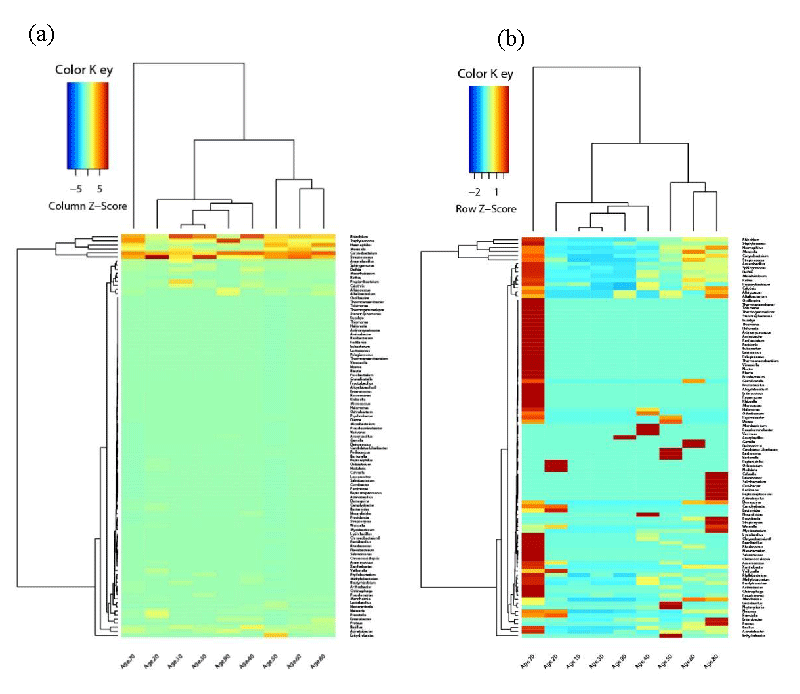
For microbiome analysis, amplification of the V3 and V4 region was confirmed as peaks around 550bp were detected via Bioanalyzer. The final study group consisted of 92 patients whose mean (+ SD) age was 67 + 16 SD years (range 19 to 93). Nasopharyngeal bacterial profiles of the patient samples were comprised of the aligned MiSeq sequence reads. As summarized in Figure 2, a total of 100 OTUs were classified up to the genus level with > 1% abundance in at least one sample. Actinobacteria, Firmicutes, and Proteobacteria dominated the bacterial profiles in the phylum level, of which Corynebacterium, Staphylococcus, Streptococcus, Haemophilus, Moraxella, Acinetobacter, and Rhizobium genera were detected with high frequency. In detail, the microbial composition of samples is summarized at the phylum, class, order, family, and genus level and portion in Figure 2. Common bacterial pathogens were present as either single pathogens or in combination with other organisms in the patient samples. These findings indicated concurrent nasopharyngeal carriage of the following pair of genera in Figure 3: Selenomonas and Rhodcoccus (correlation of 1), Anaerococcus and Rhodococcus (correlation of 0.94), Veillonella and Bacteroides (correlation of 0.97), and Veillonella and Prevotella (correlation of 0.97). In comparison, no obvious pattern of the nasopharyngeal carriage was observed between the different hospital locations, and no significant difference in the microbial community structure was observed between the male and female patients. As seen in Figure 4, Age, on the other hand, was a definitive factor in the microbiome composition for patients between the ages of over 50’s. Elderly patients included in this age group were the only ones whose nasopharynx was colonized with either Alloiococcus or Alkalibacterium. Furthermore, patients between their 60s and 70s shared a highly similar structure of microbial community at the genus level, with a visible overall pattern of pathogen incidence. For virome analysis, we were unable to obtain any viral sequence reads since the low viral concentration resulted in a failure of cluster generation on the Illumina Miseq platform.
Discussion
This prospective cohort study utilized the NGS technique to investigate the nasopharyngeal microbiota of patients hospitalized with CAP syndrome in 8 different tertiary care hospitals in South Korea. Prior to the study, negative results for 16 common respiratory viruses were confirmed for the nasopharyngeal specimens. Viruses selected for real-time PCR analysis were of interest to the researchers because of their novelty in Korea and their increasing presence in neighboring countries. Despite preparing the samples for viral enrichment, utilizing filtration, concentration, and amplification of viral nucleic acid, both real-time PCR and NGS techniques failed to detect any viruses present in the nasopharyngeal specimens. Consequently, the study was geared towards OTUs that were frequently isolated from the human nasopharynx and known as etiological agents of CAP [15-17]. The concurrent nasopharyngeal carriage of Streptococcus, Haemophilus, and Moraxella species by the patients are consistent with previous reports on the association of this particular set of bacteria with CAP. Pulmonary pathogens like Veillonella, on the other hand, have been associated with Ventilator-Associated Pneumonia (VAP) and aspiration pneumonia [18]. Veillonella is part of the normal human flora of sites such as the oral cavity and upper respiratory tract [19]. In the nasopharyngeal bacterial profiles of the pneumonia patients, Veillonella was present in combination with either Bacteroides or Prevotella. According to Falagas, et al. there has been the isolation of Bacteroides and Prevotella species from nearly all anatomic sites of infection [20]. In another study, Veillonella was also isolated along with Prevotella in VAP patients [21]. Isolation of Veillonella in combination with either of these two species, however, has not been previously reported for CAP. As previously mentioned, variations were observed in the nasopharyngeal microbiota between patients according to their age and location. The gender of the patients did not have significant implications on the bacterial community structure. The data indicate that the microbial communities of patients between the age of 60 and 70 were highly similar in composition. Furthermore, there were 2 bacterial species that were only present in elderly patients. For more detailed bacterial community structure, Alkalibacterium and Alloiococcus were only isolated from the nasopharynx of patients between the age of 50 and 90. Alkalibacterium species are lactic acid bacteria commonly found in marine organisms and salted foods like olives [22]. According to H. Liang, et al. Alkalibacterium species were among the main lactic acid bacteria isolated from paocai, Chinese fermented pickles. In Korea, kimchi, which is made of fermented cabbage, can be seen as equivalent to olives and the Chinese paocai. Although previous studies of the microbial community involved in kimchi fermentation have described lactic acid bacteria such as Lactobacillus and Lactococcus as the dominant players, results of this study indicate that Alkalibacterium may also be widespread in Korean fermented foods like kimchi [23]. The prevalence of Alkalibacterium in only elderly patients demonstrates that increased consumption of fermented foods is associated with the elderly population. Alloiococcus was another taxon present specifically in the nasopharyngeal microbiota of elderly patients. Contrary to our findings in the elderly population, previous studies have reported nasopharyngeal carriage of Alloiococcus species in children, especially in those who are prone to otitis. Although Alloiococcus has been deemed as part of the normal flora of the ear, these bacteria have also been shown to have an immune-stimulatory ability [24]. These findings indicate nasopharyngeal carriage of bacteria that typically reside in the human mouth or ear. This can be explained by the fact that the human nose is an intermediary between the ear, mouth, and lower respiratory tract, facilitating microbial colonization at these sites [25]. Rhizobium was another atypical taxon that was detected with high frequency in the nasopharyngeal specimens, across a range of patient ages and locations. Rhizobium species are plant pathogens rarely associated with human infections or identified as common residents of the human nose [26]. On one hand, organisms of the Proteobacteria phylum including Bradyrhizobium and Rhizobium species have been reported as contaminants isolated from laboratory saline solution and commercial kits for DNA isolation [27]. On the other hand, R. radiobacter is a known opportunistic human pathogen causing infections in patients with indwelling foreign material, such as catheters or prosthetics, or underlying diseases [28]. Since R. radiobacter is of the same genus, it may be possible for the Rhizobium species sequenced in this study to cause human infections, as well. Further studies should be done to test this hypothesis. Although differences in the bacterial composition existed between patients, these findings support earlier reports on the pathogen incidence associated with CAP of bacterial etiology.
Conclusion
Although the samples were obtained from patients with different general characteristics, such as age, gender, and location, the nasopharyngeal microbiota revealed common dominant species of bacterial pathogens associated with pneumonia, and respiratory infections in general.
Because samples were collected mostly from elderly patients, the viral concentration was insufficient for NGS analysis. Therefore, further investigations of the viral microbial community structure of patients with pneumonia syndrome are in progress using pediatric samples.
The large number of elderly patients present in the study group also highlights the importance of early detection of pneumonia, since increased rates of morbidity and mortality due to pneumonia are associated with the elderly population.
Our study demonstrated how NGS is a useful platform for determining the composition of nasopharyngeal microbiota. However, sufficient concentration of pathogens is required to yield sequence reads as seen in our attempts to identify any viruses in the samples.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07045711), the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MSIP) (RS-2023-00207833) and SK Bioscience (2022-I-1).
- Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, Torres A, Whitney CG; Infectious Diseases Society of America; American Thoracic Society. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007 Mar 1;44 Suppl 2(Suppl 2):S27-72. doi: 10.1086/511159. PMID: 17278083; PMCID: PMC7107997.
- (KOSTAT) SK. Cause of death statistics in 2016. Statistics Korea (KOSTAT), Government Complex-Daejeon. 2017.
- Chochua S, D'Acremont V, Hanke C, Alfa D, Shak J, Kilowoko M, Kyungu E, Kaiser L, Genton B, Klugman KP, Vidal JE. Increased Nasopharyngeal Density and Concurrent Carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis Are Associated with Pneumonia in Febrile Children. PLoS One. 2016 Dec 1;11(12):e0167725. doi: 10.1371/journal.pone.0167725. PMID: 27907156; PMCID: PMC5132320.
- Noh JY, Lim S, Song JY, Choi WS, Jeong HW, Heo JY, Lee J, Seo YB, Lee JS, Wie SH, Kim YK, Park KH, Jung SI, Kim SW, Lee SH, Lee HS, Yoon YH, Cheong HJ, Kim WJ. Interim estimates of the effectiveness of the influenza vaccine against A(H3N2) influenza in adults in South Korea, 2016-2017 season. PLoS One. 2017 May 25;12(5):e0178010. doi: 10.1371/journal.pone.0178010. PMID: 28542417; PMCID: PMC5444786.
- Lim WS, Smith DL, Wise MP, Welham SA; British Thoracic Society. British Thoracic Society community acquired pneumonia guideline and the NICE pneumonia guideline: how they fit together. Thorax. 2015 Jul;70(7):698-700. doi: 10.1136/thoraxjnl-2015-206881. Epub 2015 May 14. PMID: 25977290; PMCID: PMC4484256.
- Prevention CfDCa. Enterovirus D68 for health care professionals. October 20, 2017. https://www.cdc.gov/non-polio-enterovirus/hcp/ev-d68-hcp.html.
- Kuypers J, Campbell AP, Guthrie KA, Wright NL, Englund JA, Corey L, Boeckh M. WU and KI polyomaviruses in respiratory samples from allogeneic hematopoietic cell transplant recipients. Emerg Infect Dis. 2012 Oct;18(10):1580-8. doi: 10.3201/eid1810.120477. PMID: 23017213; PMCID: PMC3471632.
- Selvaraju SB, Nix WA, Oberste MS, Selvarangan R. Optimization of a combined human parechovirus-enterovirus real-time reverse transcription-PCR assay and evaluation of a new parechovirus 3-specific assay for cerebrospinal fluid specimen testing. J Clin Microbiol. 2013 Feb;51(2):452-8. doi: 10.1128/JCM.01982-12. Epub 2012 Nov 21. PMID: 23175256; PMCID: PMC3553926.
- Taniguchi S, Maeda K, Horimoto T, Masangkay JS, Puentespina R Jr, Alvarez J, Eres E, Cosico E, Nagata N, Egawa K, Singh H, Fukuma A, Yoshikawa T, Tani H, Fukushi S, Tsuchiaka S, Omatsu T, Mizutani T, Une Y, Yoshikawa Y, Shimojima M, Saijo M, Kyuwa S. First isolation and characterization of pteropine orthoreoviruses in fruit bats in the Philippines. Arch Virol. 2017 Jun;162(6):1529-1539. doi: 10.1007/s00705-017-3251-2. Epub 2017 Feb 11. PMID: 28190201.
- Singh H, Yoshikawa T, Kobayashi T, Fukushi S, Tani H, Taniguchi S, Fukuma A, Yang M, Sugamata M, Shimojima M, Saijo M. Rapid whole genome sequencing of Miyazaki-Bali/2007 Pteropine orthoreovirus by modified rolling circular amplification with adaptor ligation - next generation sequencing. Sci Rep. 2015 Nov 12;5:16517. doi: 10.1038/srep16517. PMID: 26558341; PMCID: PMC4642344.
- Hall RJ, Wang J, Todd AK, Bissielo AB, Yen S, Strydom H, Moore NE, Ren X, Huang QS, Carter PE, Peacey M. Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J Virol Methods. 2014 Jan;195:194-204. doi: 10.1016/j.jviromet.2013.08.035. Epub 2013 Sep 13. PMID: 24036074; PMCID: PMC7113663.
- Chun J. A metagenomic analysis of bacteria and viruses in respiratory tract samples with influenza infection. Chunlab, Inc. 2014.
- Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004 Oct;17(4):840-62, table of contents. doi: 10.1128/CMR.17.4.840-862.2004. PMID: 15489351; PMCID: PMC523561.
- Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010 Dec;38(22):e200. doi: 10.1093/nar/gkq873. Epub 2010 Sep 29. PMID: 20880993; PMCID: PMC3001100.
- Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010 Aug 1;51 Suppl 1:S81-7. doi: 10.1086/653053. Erratum in: Clin Infect Dis. 2010 Nov 1;51(9):1114. PMID: 20597676.
- Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011 Feb 1;2(1):e00245-10. doi: 10.1128/mBio.00245-10. PMID: 21285435; PMCID: PMC3031303.
- Gao Z, Kang Y, Yu J, Ren L. Human pharyngeal microbiome may play a protective role in respiratory tract infections. Genomics Proteomics Bioinformatics. 2014 Jun;12(3):144-50. doi: 10.1016/j.gpb.2014.06.001. Epub 2014 Jun 18. PMID: 24953866; PMCID: PMC4411333.
- Marik PE, Careau P. The role of anaerobes in patients with ventilator-associated pneumonia and aspiration pneumonia: a prospective study. Chest. 1999 Jan;115(1):178-83. doi: 10.1378/chest.115.1.178. PMID: 9925081.
- Shah A, Panjabi C, Nair V, Chaudhry R, Thukral SS. Veillonella as a cause of chronic anaerobic pneumonitis. Int J Infect Dis. 2008 Nov;12(6):e115-7. doi: 10.1016/j.ijid.2008.03.018. Epub 2008 Jun 10. PMID: 18547858.
- Falagas ME, Siakavellas E. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int J Antimicrob Agents. 2000 Jun;15(1):1-9. doi: 10.1016/s0924-8579(99)00164-8. PMID: 10856670.
- Doré P, Robert R, Grollier G, Rouffineau J, Lanquetot H, Charrière JM, Fauchère JL. Incidence of anaerobes in ventilator-associated pneumonia with use of a protected specimen brush. Am J Respir Crit Care Med. 1996 Apr;153(4 Pt 1):1292-8. doi: 10.1164/ajrccm.153.4.8616556. PMID: 8616556.
- Ishikawa M, Yamasato K, Kodama K, Yasuda H, Matsuyama M, Okamoto-Kainuma A, Koizumi Y. Alkalibacterium gilvum sp. nov., slightly halophilic and alkaliphilic lactic acid bacterium isolated from soft and semi-hard cheeses. Int J Syst Evol Microbiol. 2013 Apr;63(Pt 4):1471-1478. doi: 10.1099/ijs.0.042556-0. Epub 2012 Jul 27. PMID: 22843725.
- Haque MA, Lee JH, Cho KM. Endophytic bacterial diversity in Korean kimchi made of Chinese cabbage leaves and their antimicrobial activity against pathogens. Food Control 2015;56:24-33.
- Janapatla RP, Chang HJ, Hsu MH, Hsieh YC, Lin TY, Chiu CH. Nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Alloiococcus otitidis in young children in the era of pneumococcal immunization, Taiwan. Scand J Infect Dis. 2011 Dec;43(11-12):937-42. doi: 10.3109/00365548.2011.601754. Epub 2011 Sep 6. PMID: 21892897.
- Shak JR, Vidal JE, Klugman KP. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 2013 Mar;21(3):129-35. doi: 10.1016/j.tim.2012.11.005. Epub 2012 Dec 25. PMID: 23273566; PMCID: PMC3729046.
- Lai CC, Teng LJ, Hsueh PR, Yuan A, Tsai KC, Tang JL, Tien HF. Clinical and microbiological characteristics of Rhizobium radiobacter infections. Clin Infect Dis. 2004 Jan 1;38(1):149-53. doi: 10.1086/380463. Epub 2003 Dec 4. PMID: 14679463.
- Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016 May 26;8:24. doi: 10.1186/s13099-016-0103-7. PMID: 27239228; PMCID: PMC4882852.
- Chen CY, Hansen KS, Hansen LK. Rhizobium radiobacter as an opportunistic pathogen in central venous catheter-associated bloodstream infection: case report and review. J Hosp Infect. 2008 Mar;68(3):203-7. doi: 10.1016/j.jhin.2007.11.021. Epub 2008 Mar 4. PMID: 18289729.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
