Journal of Biology and Medicine
Transcriptional profiling reveals fundamental differences in iPS-derived CD34+ Cells versus adult circulating CD34+
Elmira Jalilian1,2*, William Raimes1 and Roberto Martinez Macias1
2Department of Ophthalmology and Visual Sciences, University of Illinois at Chicago, Chicago, IL, USA
Cite this as
Jalilian E, Raimes W, Macias RM (2024) Transcriptional profiling reveals fundamental differences in iPS-derived CD34+ Cells versus adult circulating CD34+. J Biol Med 8(1): 001-013. DOI: 10.17352/jbm.000041Copyright License
© 2024 Jalilian E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.CD34+ cells hold significant promise in regenerative medicine and the treatment of various vascular degenerative diseases primarily because of their ability to regenerate and differentiate into various cell types. These cells can be derived from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), or adult stem cells like blood and bone marrow, and can differentiate into endothelial, hematopoietic, myeloid, and other cell types. However, their characteristics vary based on the source, making detailed definitions essential for specific therapeutic applications. In this study, it was aimed to compare these two different CD34-positive cell populations by full genome transcriptional profiling (RNAseq). To this end, we first optimized a CD34+ cell differentiation protocol and found that Vascular Endothelial Growth Factor (VEGF) is critical for the transition of cells from mesodermal precursors to CD34+ cells. Additionally, principal component analysis (PCA) of RNAseq data showed that blood-derived CD34+ cells clustered far from iPS-derived CD34+ cells which illustrates these populations are fundamentally different. This data will be useful to better define these cell populations and facilitate the translation of regenerative approaches in this field as well as providing potentially novel diagnostic tools.
Introduction
CD34+ cells are known to be a promising stem cell source for vascular regeneration [1-5]. These cells were initially shown in human adult circulating blood cells [6] and were called “Endothelial Progenitor Cells” (EPC). These cells were later shown to be present in adult bone marrow (BM) [7-9] and peripheral blood (PB) [10]. EPCs were believed to be progenitor cells and could differentiate into endothelial cells in vitro. However, the controversy over the origin, differentiation, and cellular identity of these cells remains a potential issue [11]. Although CD34 is the main marker of EPCs [12,13], other markers such as CD133, CD31, and Tie-2 were used in combination with CD34 to more accurately identify and characterize endothelial cells and endothelial progenitor cells’ functionality, identification, and differentiation [14]. Despite a high number of studies on the clinical usefulness of CD34+ cells [15-19], it is still not completely understood what exactly these cells are and what is the exact role of this population outside of each individual specificity [20]. On one hand, a small population of circulating cells in the adult peripheral bloodstream has been shown to generate CD34+ cells [21]. It has been suggested that these cells can be further stimulated to migrate, proliferate, or differentiate into a more mature lineage and are able to either directly [22-25] contribute or indirectly [26-28] support, vascular regeneration [29]. On the other hand, during embryonic development, endothelial cells develop from a precursor population [30]. These “true” embryonic progenitor cells also express CD34, which are which are also shown to participate in vessel regeneration [31]. These embryonic-derived CD34+ cells are most likely to be very different from adult peripheral blood CD34+ cells. To date, it is not well understood what these differences are [32]. Therefore, comparative studies of different CD34+ cell populations based on their broad range of molecular characteristics may contribute to understanding the difference between all these populations and would offer an insight into overlapping properties of the cells that express CD34. In this study, we intended to use gene expression profiling to better characterise the identity of circulating adult CD34+ and iPS-derived CD34+. Thus, we first aim to establish an efficient protocol to generate iPS-derived CD34+ cells in sufficient quantity in vitro and then compare their gene expression profiling with Adult CD34+ cells isolated from cord blood and peripheral blood.
Results and discussion
in vitro derivation of CD34+ cells from induced Pluripotent Stem Cells (iPSCs)
In order to establish a protocol to generate a high yield of CD34+ cells in vitro, sequential experiments were developed (Table 1). Key parameters such as factors, concentrations, and time course of factors and substrates were modified in further continuous experiments to optimize the best conditions in which to grow CD34+ cells. Each experiment provided continuous feedback which was used for the further optimization process. At the end of the experimental process cells were fixed with 4% PFA and then processed for immune-staining to assess the CD34 expression. Outcomes from the first set of experiments were used to change the design for the next experiment. Conditions with more cell death and no CD34 expression were excluded. Better conditions were selected to repeat in the next experiments in addition to new conditions. In all experiments, each treatment condition was performed in triplicate. Relative expression of CD34 was analyzed in a semi-quantitative score method explained in method 1.6. Briefly, the relative expression of CD34 was assessed as relative CD34+ cell yield (number sign), cell detachment (low/ medium/ high), and CD34+ coverage (dispersed/ aggregated / extensive) for each treatment condition.
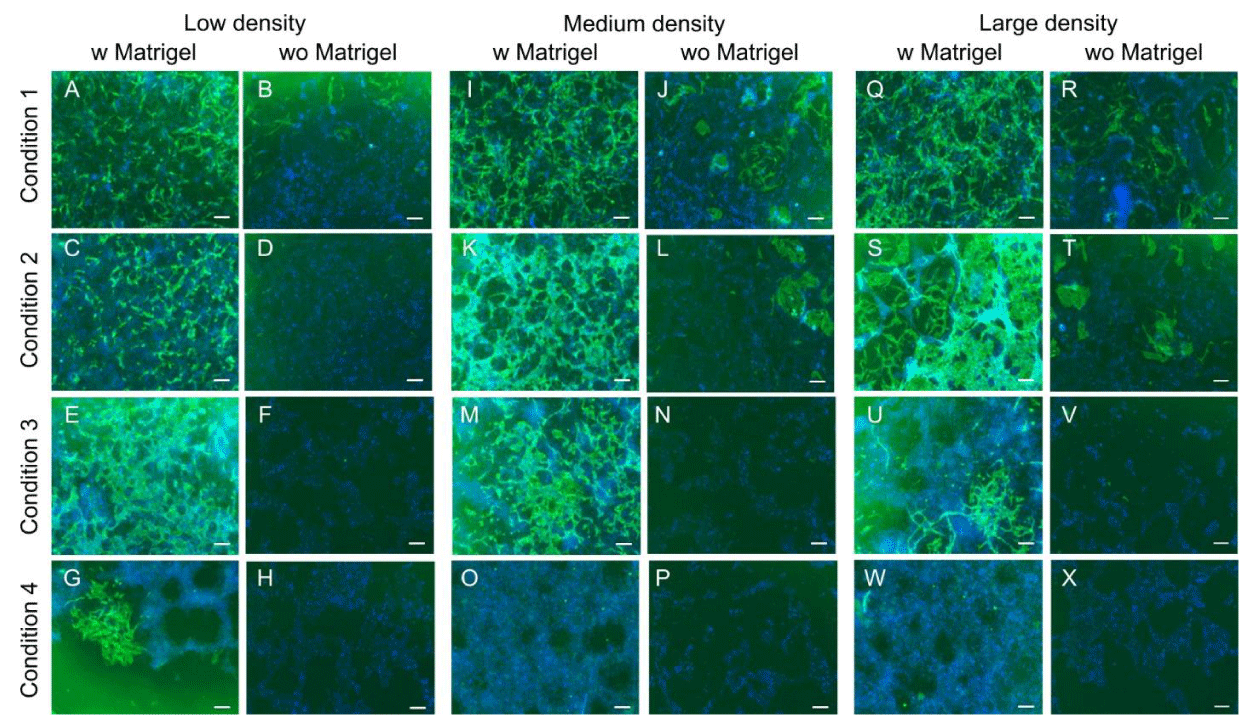
The first step of optimization was to find the most suitable medium. Six different media were tested with different combinations of factors in 96 well plates including the media used in two papers [33,34]. Among all the media tested, DMEM/F12 in combination with B2+ N27 was the best medium to induce the most CD34 positive cells and it was selected for the remainder of the experiments. Data shown in (Supplementary Figure 1). Over the optimization procedures the concentrations of Activin A 25 ng/ml, VEGF 50 ng/ml and bFGF 25ng/ml were not changed. BIO and SB431512 were used in the concentration of 1.5 µM and 10 µM initially. However, conditions having BIO 1.5 µM had a big area of cell death presumably because of the high concentration of BIO (Supplementary Figure 2). Therefore, the concentration of BIO was further reduced to 0.5 µM and finally adjusted at a final concentration of 0.15 µM. Furthermore, conditions containing SB 431542 10 µM from day 2, also showed a high amount of cell death. Therefore, in further experiments, the concentration of SB 431542 was reduced to 2 µM (Supplementary Figure3). After optimization of the medium and concentrations of factors, since enough CD34 enrichment was not observed an attempt was made to optimize the protocols by modifying different parameters such as substrate and different combinations of factors. Furthermore, since variable results were observed over repeated experiments up to this point, it was assumed that the number of cells initially plated might also be an effective factor in the induction of CD34+ cells. Therefore, in the next experiment substrate was modified in parallel with three different cell numbers (low, medium, and high) to find out its influence on enhancing iPS-derived CD34+ cells. Furthermore, to take advantage of the 3D culture system and create a more physiological microenvironment, extra Matrigel was added on top of the medium and then compared with the 2D culture condition. iPSC cells were passaged and plated in high density (about 40,000 cells), middle density (25000 cells), and low density (13000 cells) in 96 well plates coated with Matrigel. Four different conditions were tested. to create 3D culture conditions additional 2.5% Matrigel (1:80 final working ratios) was mixed into the medium and added to the plates and similar conditions were compared to 2D culture conditions (Table 1). Immunostaining results showed that adding extra Matrigel (2.5%) on top of the cells could dramatically increase the CD34 yield in all conditions (Figure 1).
Furthermore, the highest expression of CD34 was observed in middle and high density. Conditions 1 and 2 had the highest expression of CD34 in middle and high density in the 3D culture system whereas conditions 3 and 4 had the highest expression in low and middle density (Error! Reference source not found.). Therefore, it was concluded seeding cells in high density (40000 cells/well) and 3D culture condition is sufficient to induce the most CD34 positive cells. Moreover, consistent results using bFGF in the treatments were not observed in repeated experiments. Since CD34 expression was observed in conditions without bFGF, this factor was excluded from later experiments.
In the further optimization process, the effect of BMP4 and BIO were assessed at different time points and in different combinations (Supplementary Figure 4) [35-37]. Immunocytochemistry results showed that BMP4 for three days is more effective than one day in inducing CD34 expression. Furthermore, conditions containing BMP4 from day 1 to 5 alone or in combination with other factors did not show any expression of CD34 which could indicate that BMP4 is a critical signaling molecule required for inducing the mesodermal lineage only in the very early phase of mesodermal induction. However, having BMP4 alone is not effective and BMP4 should be applied in combination with other factors.
Next, the effect of BMP4 alone, condition 10 in (Supplementary Figure 4) or in combination with different factors was assessed for different time points and it was compared with the condition without BMP4. It was intended to find out if BMP4 would be sufficient enough to induce the high amount of CD34 or if adding other factors to the cocktail is important to induce the CD34 expression. Condition with only VEGF treatment was also considered, condition 11 in (Supplementary Figure 4) to assess if VEGF alone would be enough to induce CD34 expression. Previous experiments could clearly illustrate that a low concentration of SB 431542 (2 µM) had much lower cell death compared to high concentrations (10 µM) which was again confirmed in the current experiment. In the current experiment, SB 431542 (2 µM) was also used at different time points to find out its effect in increasing the CD34 expression and comparing this to conditions without SB 431542. Furthermore, the effect of BIO for the first 1 or 3 days was also assessed in combination with other factors, all conditions are shown in (Supplementary Figure 4). Similar to previous experiments, cells were seeded in triplicate onto Matrigel-coated 96-well plates in a 3D culture system at 4 × 104 cells/well. Experimental timelines ran 6 days long and consisted of growth factors addition in fresh medium on days 1, 2, and 4. Cells were fixed and stained on day 6.
Immunocytochemistry results showed that BMP4 for three days is more effective than one day in inducing CD34 expression. However, having BMP4 alone, condition 10 (Supplementary Figure 4) is not effective and BMP4 should be applied in combination with specific factors. From the current experiment, it could be concluded that BIO is a critical factor to be considered with BMP4 in the cocktail. Conditions containing BMP4 but not having BIO had no or very low expression of CD34 (conditions 1, 2, 3, 10, and 11). The presence of BIO either for one or three days seems to have a high influence in inducing the CD34-positive cells (conditions 5, 6, and 7). However, it is not clear how would be the influence of BIO if it would be kept longer up to day 5. Therefore, this hypothesis was assessed in the next experiment. Furthermore, SB 431542 was another important factor to be considered albeit in combination with specific factors and specific time periods. Conditions without SB 431542 had very low or no expression of CD34, conditions 1, 2, 10, and 11 (Supplementary Figure 4) which could show the importance of this factor in CD34 induction. Moreover, conditions with the highest expression of CD34 had SB 431542 only for two days. Conditions containing SB 431542 for more than 2 days (from day 2 to 6) had a big area of cell death and no or very low expression of CD34, conditions 3, 4, 8, and 9. This observation could suggest that SB 431542 only for the last two days is sufficient enough to induce the most CD34-positive cells. This was consistent with previous findings showing that inhibition of TGF-ß before mesodermal induction results in a reduction of CD34 expression [38]. Therefore, SB 431542 was considered to apply for only two days during the late phase of treatment (days three to five). Moreover, in conditions containing BMP4 from day 1 to 5 alone, conditions 10 or in combination with other factors, condition 1 did not show any expression of CD34 which could indicate that BMP4 is a critical signaling molecule required for inducing the mesodermal lineage only in the very early phase of mesodermal induction. Additionally, having VEGF (50 ng/ml) alone was not enough to induce the CD34 expression, condition 11. This experiment could show the importance of BIO and BMP4 in CD34 induction. Since this experiment could show the importance of BIO in CD34 induction, the next experiment was designed to further investigate the longer effect of BIO and its effect in combination with or without BMP4 and SB 431542, all data shown in (Supplementary Figure 4).
From the fact that applying BIO either for 1 or 3 days was shown to be very effective in inducing the high number of CD34, a further experiment was designed to find out the longer effect of BIO factor in the absence or presentence of SB 431542 or BMP4 in CD34 induction. BIO was considered for all conditions for either 1, 2, or 5 days. Furthermore, BMP4 was also considered for either 1 or 3 days which was combined with SB 431542 in some of the conditions to compare the different combinations of factors. Furthermore, some conditions evaluated the effect of BIO in the absence of BMP4 with or without SB 431542 (all conditions shown in supplementary data 5). Immunocytochemistry results revealed that conditions adding BIO to the medium for a longer time period (day zero to five) suppressed the expression of CD34-positive cells, conditions 3 and 4 in (Supplementary Figure 5).
BIO is necessary only for the early phase of treatment but becomes detrimental at later stages. Furthermore, the combination of this factor with SB 431542 (condition 3 supplementary data 5) from day 3 to 5 had a relatively bigger area of cell detachment compared to the condition without SB 431542. Conditions having BIO and BMP4 for 3 days and then VEGF + SB 431542 for 2 days had comparatively more CD34 expression compared to similar conditions without SB 431542, Condition 5 vs. 6. Furthermore, it was observed that conditions having BIO for 3 days but excluding BMP4 from day 2 conditions 5 & 6 still had high expression of CD34 and it was comparatively more if SB 431542 was added to the cocktail in the last 2 days. However, if BIO was excluded from day 2 and instead BMP4 was included up to day 3 conditions 7 and 8 (Supplementary Figure 5), less CD34 was observed compared to 3 days of stimulation with BIO. Similarly, SB 431542 had slightly higher CD34 coverage compared to the VEGF-only condition in the last 2 days of treatment. In conditions 9 and 10 BMP4 is excluded from day 1 and is only included in condition 10 from day 2 to 5. A comparison of the two conditions shows that including BMP4 is effective in increasing the number of CD34-positive cells. Comparisons of conditions 9 and 11 supplementary data 5 which are similar and the only difference is the presence or absence of SB 431542 emphasize the positive effect of SB 431542 in inducing the CD34 positive cells. Furthermore, excluding all factors except VEGF from day 2 does not seem to be effective in CD34 induction (condition 12). Condition without any of these factors does not show any CD34 expression condition 13. Comparing all possible factor compositions shows that including BIO and BMP4 together for three days is more effective than having each of them separately for 3 days or including one of them for 1 day and the other for 3 days. Furthermore, including SB 431542 for the last two days is effective for increasing the number of CD34-positive cells. This was consistent with previous findings showing that inhibition of TGF-ß before mesodermal induction results in a reduction of CD34 expression [38]. Moreover, in all current experiments, Activin A was removed from day 2. Comparing immune-staining results from this experiment with the previous experiment which contained Activin A for three days reveals that the presence of Activin A for three days is more effective than 1 day. Therefore, up to this point, combining Activin A, BIO, BMP4, and VEGF for the first day and then SB 431542 and VEGF only for two days seems to be very effective to induce the greatest number of CD34 positive cells (Supplementary. Figure 5).
VEGF is critical for the transition of cells from mesodermal lineage into embryonic-derived CD34+ cells
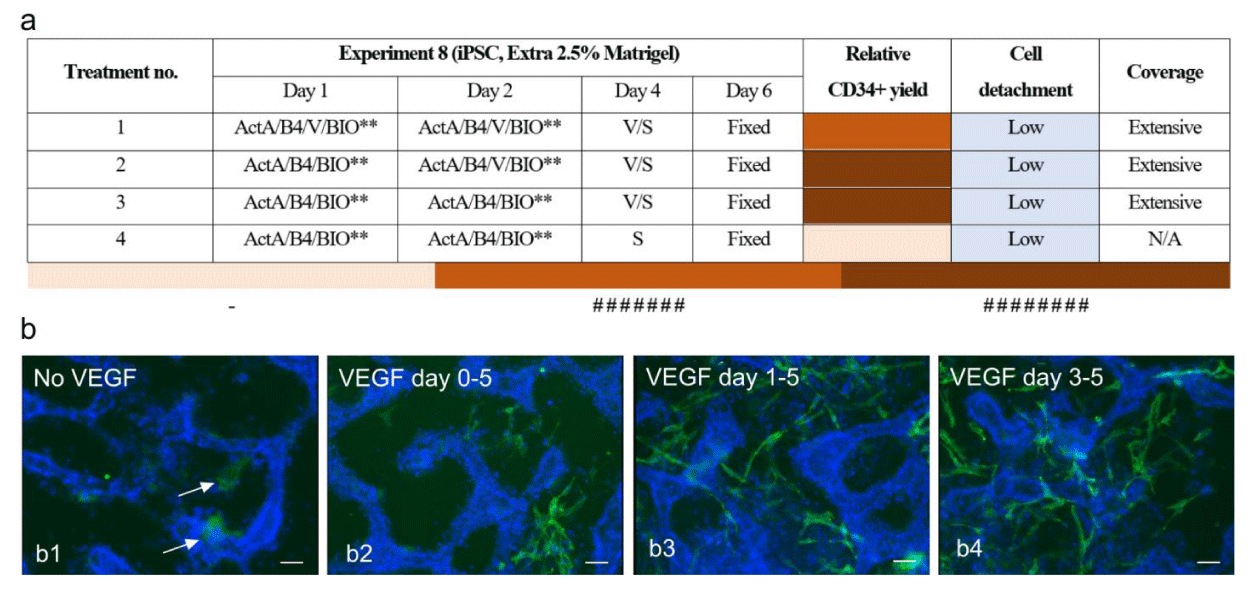
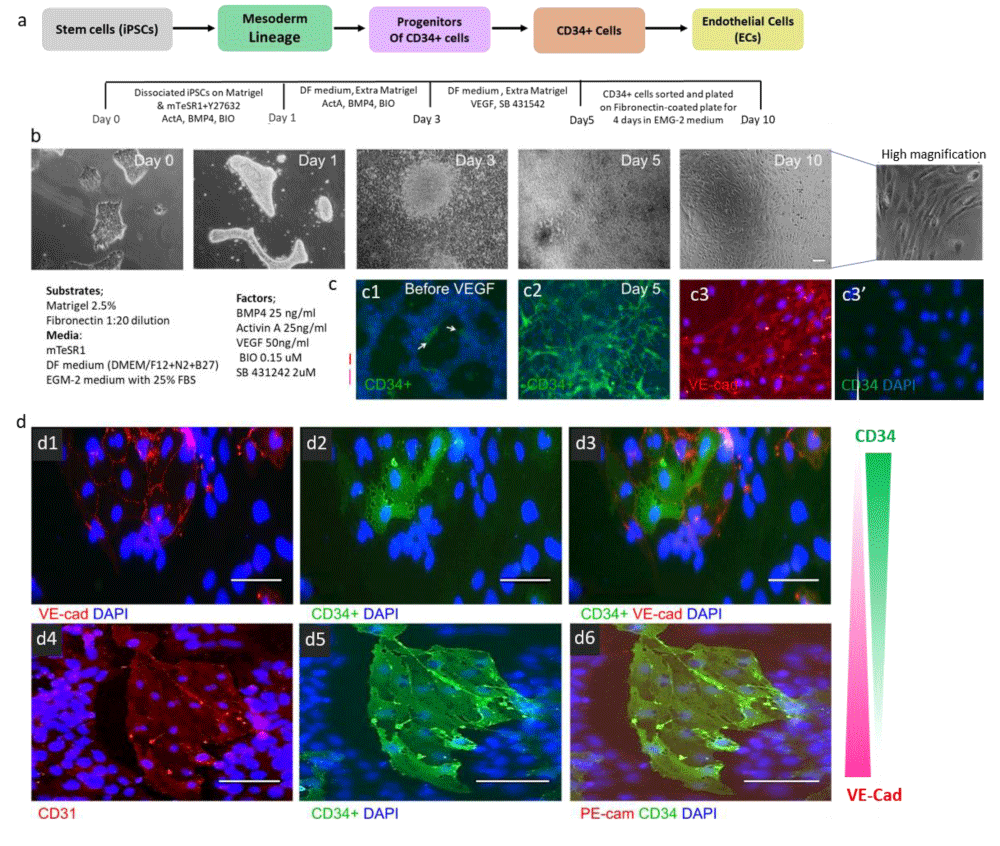
VEGF signaling has been demonstrated to be critical for the differentiation of CD34+ cells throughout embryonic development, as evidenced in several studies. It has also been incorporated into various differentiation protocols, typically from day zero until day 5 of the process [39-41]. Comparing different conditions with or without VEGF over different experiments showed that VEGF is the key factor for the generation of CD34+ cells and no expression was detected without VEGF. So far, we and others used VEGF right from the beginning for all differentiation protocols. However, up to now, no one has clearly shown in which specific stage of differentiation VEGF is crucial to generate progenitors of endothelial cells. Therefore, in the current study, we intended to find out if VEGF is needed from day zero up to day five. Or if excluded from different time points during the treatment, whether it would still affect the induction of CD34+ cells. Treatment conditions are shown in the table in (Figure 2). This experiment was repeated three times and similar results were observed from both experiments. (n = 3 biological replicate and n = 9 technical replicate). We found that adding VEGF only during the late phase of treatment and for only two days was sufficient to induce the expression of CD34-positive cells. Interestingly, Immunocytochemistry results of cells just before adding VEGF at day three, showed faint CD34 expression, arrows in NO VEGF condition in (Figure 2).
To sum up, the final optimization illustrated that the administration of BMP4, Activin A, and BIO (Wnt signaling activator) at an early phase induces the cells through the mesoderm lineage. Further stimulation of cells at this stage (Figure 2c1) with VEGF and SB431542 (TGFβ-receptor type one inhibitor) during the second phase of treatment leads to efficient differentiation of CD34+ cells from human iPSCs within five days (Error! Reference source not found. c2). Immunocytochemistry analysis of CD34+ cells in this stage illustrated that CD34 and VE-Cadherin expression was inversely related (Figure 3 d1-d3). This suggested that CD34-expressing cells generated with our protocol were more at the immature stage of progenitor cells and had not completely differentiated to ECs and were indeed progenitors of endothelial cells. Furthermore, CD31 and CD34 appeared to be co-localised suggesting that they were vascular endothelial cells (Figure 3 d4-d6). CD34+ cells were purified with CD34-labeled beads using the MACS technique at day 5 of differentiation. CD34+ cells were plated on Fibronectin-coated plates in EGM-2+ 25% serum containing VEGF 165 (50 ng/ml) and SB 431542 (2 µM) for extra 4 days. Bright Field images of isolated cells after 4 days in culture show spindle-like cell morphology, indicating a mature endothelial cell phenotype (Error! Reference source not found. b day 10). Furthermore, immunocytochemistry of these cells illustrated that the vast majority of the isolated cells express the endothelial cell marker VE-Cadherin, whereas CD34 expression has been strongly reduced (Figure 3 c3 & c3’). This confirmed the endothelial nature of isolated cells and further differentiation of them towards endothelial cells.
It is important to mention that using two iPS batches from the same donor, we found strong CD34-expressing cells appeared in conditions containing only SB 431542 and no VEGF. Further observations revealed that against similar cell numbers seeded in the first place, the cell density in the second set of experiments was much higher after five-day treatment compared to the first set of iPS cells. Therefore, it was assumed that because of high confluency and a lack of oxygen, cells might have generated their own VEGF at this stage to compensate for their environmental conditions. To test for this, we used aflibercept (Eylea), a vascular endothelial growth factor (VEGF) inhibitor (1:100 dilutions) which is an anti-VEGF drug, and cells were treated based on the protocol established earlier. On day three of differentiation, Elyea was added in combination with SB 431542 and was maintained until day five. Then cells were fixed on day five for further immunostaining analysis. Immunostaining results illustrated that in the presence of Eylea, the expression of CD34 was highly suppressed (very dim expression) (Supplementary. Figure 7). Using Eylea resulted in the generation of a homologous population of cells with no indication of CD34 expression. Thus, for our further RNA-sequencing experiments, we used Eylea (VEGF-blocker) to suppress all CD34 expression between days three to five to have more homologous population for non-expressing CD34 positive cells and compared this population with iPS-derived CD34+ cells exposed to VEGF.
Transcriptomic comparison of iPSC-derived CD34+ cells versus cord blood and peripheral blood
In order to establish how CD34+ populations from different sources were related to each other, we applied a genome-wide transcriptional profile. To this end, the Truseq Illumine RNA sequencing platform was used. For the adult CD34+ cell population in this study, we used commercially obtained CD34+ cells that have been purified from adult peripheral blood and cord blood. Since previous studies illustrated that macrophages/monocytes share lineage traits with CD34 epithelial progenitor cells [42] and cells isolated from macrophages/monocytes could be involved in blood vessel regeneration by secreting angiogenic growth factors [43] we also included a sample of CD14+ monocytes to compare its genes transcriptional profiling with the rest of the cell populations. For the iPSC-derived CD34+ cell population, we used our protocol to differentiate iPSC towards the EC lineage in the presence of VEGF or a VEGF blocker. FACS was then used to purify the CD34+ cells from the cultures that contained VEGF. In the case of the cultures that contained the VEGF-blocker, all cells were used without purification. RNA was then isolated from all eight sample populations. Trizol extracted RNA from all samples was then sent to the sequencing facility to Institute of Child Health at University College London (UCL) be processed for RNA-sequencing. Agilent RNA Integrity Number (RIN) was first used to assess RNA quality for RNA sequencing analysis. Representative RIN is shown in (Supplementary Figure 7). After quality control, RNA samples were further proceeded for library preparation and sequencing on the Illumina TruSeq RNA v2 platform. RNAseq data were further normalised by one of our collaborators with bioinformatics expertise (Monte Radeke, UCSB, Santa Barbara, USA). Our RNAseq data was normalised using the TMM method that was applied in the edgeR Bioconductor package (version 2.4.0). The number of counts from RNAseq data were adjusted to reads per million gene alignments per 1kb transcript length (CPMK) to facilitate transparent comparison of transcript levels both within and between samples.
Validation of RNAseq data
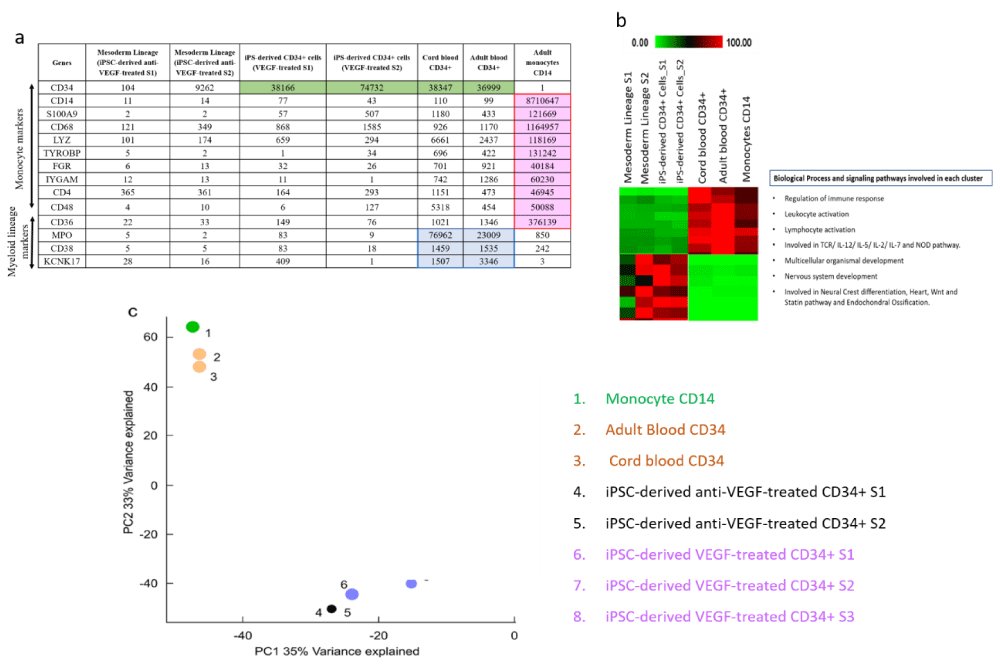
Once the RNAseq raw data was processed and turned into an Excel spreadsheet with 13 columns (for each sample) and around 16000 lines (for different genes), we first checked the different sample populations expressed specific genes we expected them to express. Since CD14 expression was the selection criteria for CD14+ monocytes, we anticipated that gene to be highly expressed in this population. The same applies to CD34 in CD34+ cell populations. As shown in CD14 was strongly expressed in CD14 monocytes while its expression was very low in the rest of the cell populations. Furthermore, CD34 expression was high in all CD34+ populations lined with green and low in CD34- samples, confirming the efficiency of the cell sorting used to purify these samples. The monocyte population is anticipated to strongly express monocyte markers such as S100A9, CD68, LYZ, TYPOBP, FGR, ITGAM, CD4, CD48, and CD36 lined with red. These genes were highly expressed in the CD14 population, which was another confirmation of the validity of the RNAseq data. Furthermore, classic markers of the myeloid lineage (MPO, CD38, and KCNK17) were highly expressed in blood-derived CD34+ cells lined in blue (Error! Reference source not found. a). These comprehensive analyses not only confirmed the expected gene expression patterns within respective cell populations but also provided insights into the validity of the RNAseq data and the efficacy of the experimental procedures utilized.
After the validation of RNAseq data by choosing selection criteria, genes were further investigated within the groups to get more insight into each specific group. AutoSOME is used to create clusters according to the similarity in expression between different groups. Outcome results illustrated that the three blood-derived samples including cord blood CD34+, adult blood CD34+ and CD14 monocytes formed the largest cluster (Error! Reference source not found. b). The next biggest cluster contained highly expressed genes in iPS-derived CD34+ (VEGF-treated) and iPSC-derived (before VEGF-treatment). These data confirmed that cell populations from the same origin tend to have similar levels of gene expression. For further analysis of these clusters, DAVID online tools were used to find whether there were any biological processes enriched within each cluster [44]. David showed the involvement of the first gene cluster in the regulation of immune response, leukocyte and lymphocyte activation. This is not surprising since these cells are all part of the immune system. However, analyzing the other clusters showed their involvement in very different biological pathways. Because AutoSOME [45], did not show how closely related or how far apart the different cell populations are from each other, Principle Component Analysis (PCA) was used in the next step [46]. PCA was performed by using an “R” software module. Then, a plot was generated to visualize the specific gene signatures that represent the similarity between the sample populations. The 8 samples were clustered roughly in 2 different groups. All blood-derived cells (cord/adult CD34+ and CD14+ cells) were clustered together (labeled 1, 2 &3) which were clearly segregated from iPS-derived CD34+ and CD34- cells (labeled 4, 5, 6, 7 &8) (Error! Reference source not found. c). This data surprisingly demonstrates that adult CD34+ cells are fundamentally different cell populations from embryonic-derived CD34+ cells. This data is also consistent with AutoSOME data which clustered all blood-derived cells together and separated from iPS-derived CD34+ cells. This data suggests that the origin of derived cells (embryonic versus adult) is more important than the expression of specific markers to indicate the similarities between different cell populations.
Discussion
In this study, the main aim was to conduct a gene expression analysis to better understand the nature of CD34+ cells from different embryonic and adult origins. In order to achieve this, we first needed to develop an efficient protocol to derive CD34+ cells in larger quantities. Therefore, in the first part, we optimized a protocol to reproducibly differentiate iPS cells into CD34 endothelial progenitor cells, which enabled us to isolate these cells in sufficient quantities. This study showed that initial mesodermal commitment was achieved using a cocktail of BMP4, Activin A, and BIO in serum-free culture conditions for three days and further EC lineage commitment was achieved with VEGF and SB431542 (Alk5 inhibitor) for two days. Previous studies have shown the importance of BIO (WNT signaling activator) in the presence of Activin A (as a part of TGF-ß) in a very early phase of mesodermal induction [47-49]. The current research also found the presence of these two factors to be crucial in the early phase. Although, BMP4 has been shown to play a significant role in the differentiation of stem cells towards a mesodermal lineage [37,50] in our hands administration of BIO and Activin A with or without BMP4 resulted in high expression of CD34. This suggests that BMP4 is not mandatory for the early phase of mesodermal induction. However, the cluster-like morphology of CD34 expressing cells in the presence of BMP4 (versus singly dispersed cells in the absence of BMP4) BMP4 might influence migration and proliferation of already committed precursors rather than their determination.
In contrast, the crucial role of VEGF in endothelial lineage commitment is well-known [51,52]. However, our finding that VEGF is not needed during the early phase of differentiation is less well-known. In fact, most investigations aiming to derive ECs from stem cells add VEGF right from the beginning of their differentiation protocols. Although some previous studies suggested that VEGF likely regulates the survival or propagation of CD34 progenitor cells, and not necessarily their differentiation [33,34], our studies could clearly show that VEGF plays a key role in the transition of precursors to CD34 progenitor cells. Generated CD34+ cells exhibited strong expression of this marker after 5 days of differentiation treatment. CD34 and VE-Cadherin expression were inversely related at this stage, which indicates our CD34+ cells are progenitors of ECs. Additionally, further culture of purified CD34+ cells on fibronectin differentiated them into completely mature ECs after 4 days. These cells formed a homologous monolayer with a cobblestone appearance that exhibited strong expression of VE-Cadherin and confirmed the endothelial nature of our isolated cells.
In our study, we observed a significant increase in CD34+ cell proliferation when an additional layer of Matrigel was applied to the cultures. This enhancement is likely due to the improved three-dimensional matrix provided by Matrigel, which more closely mimics the natural cellular microenvironment. Matrigel’s complex composition, resembling tissue extracellular matrix, facilitates better cell-cell interactions, and signaling, and potentially influences mechanical aspects like stiffness and porosity. These factors collectively contribute to an environment that promotes cell survival, proliferation, and differentiation [53,54].
Overall, this protocol was shown to be a very efficient method, which was time-effective, involved fewer steps compared to other published protocols, required less cell manipulation, and was reproducible over repeated experiments. It was demonstrated that at the end of this protocol, most of the cells were expressing the CD34 marker. Generated CD34+ cells exhibited strong expression of CD34 after five days of differentiation. Isolation of the CD34+ cells (by MACS) clearly demonstrated that these cells are committed to the EC lineage as they expressed several EC markers.
Two iPS cell batches used in this work were generated by two different lab members but from one donor (BJ iPS cell line). However, besides the similarity of the protocol and the source of the somatic cell line (BJ skin fibroblast) that was used to generate the iPS cells, it appeared that iPS cells behaved slightly differently from batch to batch in their proliferation rate and further response to specific factors such as “SB 431542 only condition” during the differentiation protocol. This could demonstrate the sensitivity of each iPS batch and indicate how the confluency of the cells can affect their response to various growth factors which is essential for directing the differentiation process from early mesendoderm via mesoderm towards a more differentiated CD34 progenitor cells. The sensitivity of iPS cells in response to stimulus factors was also evident within one cell line but with different passage numbers. It was observed that as the passage number was increasing the capacity of iPS cells to differentiate into CD34 positive cells was reducing and this was noticeably evident after passage number above 22. The limitations of the cell lines at high passage numbers should be taken into consideration, especially in the level of cell therapy products for therapeutic applications.
Finally, Overall, RNAseq data revealing a distinct transcriptome profile in different CD34+ / CD34- populations, suggested that a five-day differentiation protocol profoundly affected iPSC cells, to differentiate them into endothelial lineage which was reflected in their gene expression levels. After the validation of RNAseq data, by choosing selection criteria, genes were further investigated within the groups to get more insight into each specific group. Our investigations showed high expression levels of monocyte/macrophage markers in CD14+ cells which is not surprising as is known these cells are a part of the immune system. However, some of these genes (TYROBP, FCER1G, HLA-DRA, S100A9, and ITGB2), were also shown to be expressed in so-called “early epithelial progenitor cells” [55,56]. Our investigations displayed the high expression levels of these genes in the CD14+ cell population. Therefore, this data confirms the validity of previous studies regarding the characteristics of early epithelial progenitor cells, and that they share lineage traits with immune cells, specifically macrophages and monocytes
PCA analysis to decompose the overall variability of gene expression data indicated that iPS-derived epithelial progenitor cells and iPS mesoderm (with anti-VEGF) had the greatest similarity in their gene expression levels. This should be due to the fact that these samples came from the same cell sources (+/- VEGF for two days). However, comparing VEGF versus anti-VEGF treated populations, suggests that adding VEGF is sufficient to change the gene expression profile that could be completely distinguishable in an overall gene expression analysis. This could also confirm our in vitro experiments which showed high expression of CD34 cells in the VEGF-treated population. Furthermore, PCA analysis grouped cord and blood-derived CD34+ cells together with a small distance from CD14 monocytes which indicates reasonably high similarity between these populations and suggests that all blood-derived cells are closely related and come from the same origin.
However, it was surprising to find that cord and peripheral blood-derived CD34+ cells were clustered far from the iPSCs. This could suggest that blood-derived CD34+ cells are not closely related to mesodermal cell types against what was assumed before and therefore, may have some different stemness properties. Revealing differences in gene expression patterns between embryonic and adult progenitor cells provides a molecular basis for the discrepancies in the efficiency of different clinical trials and highlights the importance of a detailed definition of these cell types used in clinical trials. A better understanding of these cells will reduce the challenges in isolating a heterogeneous population of cells and will help to enhance the potential clinical benefits of CD34 cell-based therapy.
Further, in vivo, experiments would be interesting to find out if injecting iPS-derived CD34+ cells into damaged vasculature will be useful to see whether these cells can integrate into vasculature and differentiate into endothelial cells. In summary, our protocol could be used as a platform to develop CD34+ cells into more reliable therapeutic products, and our RNA-seq data facilitates a better understanding of the origin of different CD34+ populations which will facilitate the translation of regenerative approaches in this field.
Methods
Human induced pluripotent cell culture
BJ iPS cell lines were reprogrammed from human fibroblasts by two independent lab members at UCL Institute of Ophthalmology. iPSCs used between passage number 10 to 25. Cells were grown on Matrigel-coated plates (diluted 1:15 in Knockout DMEM and the final concentration of 1:30) in mTeSR1 medium (STEMCELL Technologies). Routine culture maintenance for BJ iPS cells was prepared when cells were over 70% confluent. Cells were washed with PBS and incubated in Cell Dissociation Reagent (Stem cell technologies) for 2 to 4 minutes at room temperature (RT). Cells were checked every minute under the microscope until the edge of the colonies started to curl. Then Reagent was aspirated and 6ml mTeSR1 was added. Cells were scrapped and split in 1:3 ratios in a total of 6 ml mTeSR1. Media needs to be changed every day with fresh media.
Differentiation protocol to generate iPS-derived CD34+ cells
Confluent iPSCs dissociated by gentle enzymatic treatment 1X TrypLE (Life technology) for 7 minutes at RT. Cells were then diluted with mTeSR1 and centrifuged at 800 g, for 3 minutes at 25 °C. Cells were re-suspended in 5ml mTeSR1 and Rock inhibitor (Y276332 (Calbiochem). Then cells were counted and seeded onto Matrigel-coated 96 plates at 4 × 104 cells/well and left in an incubator for 48 hrs. Differentiation was induced two days after by replacing mTeSR1 with differentiation media (DF) (DMEM/F12 + B2 + N27) and timed addition of the following growth factors: 25 ng/ml Activin A (Peprotech, 120-14), 30 ng/ml (BMP) 4 (Peprotech, AF-120-05) and 0.15 µM BIO (TOCRIS, 3194) with extra Matrigel 1:80 final ratio. This medium was refreshed the next day with the same factors. Then factors were replaced to 50 ng/mlVEGF165 (Peprotech, 100-20) and 2 µM SB 431542 (TOCRIS, 1614) from day 3 up to day 5. Cells were fixed at day 5 and immunocytochemistry for CD34 was performed to identify the cells of interest.’ The CD14+ Monocytes (Catalog #: 4Y-125A), Human Cord Blood CD34+ (Catalog #: 2C-101), and human CD34 peripheral blood (Catalog #: 3Y-101C) cells were obtained from Lonza. RNA isolation was then performed on these cells for subsequent sequencing analysis.
Immunocytochemistry
Cells were washed with PBS and fixed with 4% paraformaldehyde (PFA) for 10-15 minutes at room temperature (RT). Then cells were washed two times with PBS and then incubated with blocking buffer (blocking buffer: 1% BSA, 0.1% Triton, 0.01% tween 20/PBS.) for 30 minutes. After that, primary antibodies were diluted into a blocking buffer and were added per well plates. Primary antibodies were incubated for 1 hour at RT. Then cells were washed three times with PBS (5 minutes each). Then secondary antibodies IgG (Alexa Fluor 488, 594 Goat anti-mouse, Invitrogen) all were diluted 1/200 in blocking buffer and were added to the wells. Incubation time for secondary antibodies was 1 hr. After incubation time, cells were washed once with PBS for 5 minutes. Then 1 µg/ml dilution of Hoechst was added for nuclei staining only for 30 seconds. Cells were then washed for another 5 minutes with PBS and then were examined under the fluorescent microscope.
Microscopy
For all light imaging microscopy, a photomicroscope (Zeicc Axiophot) was used which was attached to a CCD camera (ORCA-ER (Hamamatsu). For each staining, pictures (at least 4 pictures from each plate) were taken with different magnifications in an attempt to represent most faithfully. For all Fluorescence microscopies, we used upright Axioscope, and inverted S100 fluorescence microscopies (Carl Zeiss) were used to analyze the immune-staining results.
Semi-quantitative method to score the expression of CD34
Regarding the scoring method using “number sign” conditions with 1 or 2 small clusters or few dispersed cells were scored with 1 number sign (#), whereas conditions with 2 to 5 small clusters and/or a few numbers of dispersed CD34 positive cells covering around 5% - 10% of the surface were scored with 2 number sign (# #). Conditions having between 5 to 10 clusters and/or quite high number of dispersed cells covering around 15% - 20% of the surface were scored with 3 number signs (# # #). Conditions having 10 to 20 small and medium clusters with a high number of dispersed CD34 expressing cells covering between 20% - 30% of the surface were scored with 4 number signs (# # # #). Furthermore, conditions with a considerable number of small/medium clusters and a high number of dispersed cells covering between 30-40% of the surface were scored with 5 number signs (# # # # #). Similar conditions with comparatively more coverage of CD34 expressing cells between 40% - 50% of the surface were scored with 6 number signs (# # # # # #). Conditions having a very strong expression of CD34 covering between 50-60% of the surface containing big clusters of CD34 expression and a very high number of dispersed cells were scored with 7 number sign (# # # # # # #) whereas comparatively higher percentage coverage between 60% - 70% was scored with 8 number sign (# # # # # # # #). The relative expression of CD34 was analyzed in a semi-quantitative score method. Relative CD34+ cell yield (number sign), cell detachment (low/ medium/ high), and CD34+ coverage (disperse/ aggregated / extensive) were qualitatively assessed for each treatment condition by two independent observers. Since in some conditions, a big region of cells was dead and detached from the surface, cell detachment was also scored in a semi-quantitative way according to three low, medium, and high levels of detachment. Furthermore, it was found that CD34-expressing cells in different conditions had different morphologies. Some conditions had more cluster-like structures whereas others were dispersed cells expressing CD34. Therefore, conditions were also categorized as either dispersed, aggregated, or in the case of having a strong expression of both categorized as extensive. All experiments with no expression of CD34 were shown by a minus sign (-).
Fluorescence-activated cell sorting (FACS)
MoFlo XDP (Beckman Coulter) was used to sort different unfixed samples of iPSCs. Cells were labeled with fluorescence-conjugated antibodies. FACS buffer was composed of PBS (pH 7.2), 1% BSA, and 2 mM EDTA. The whole process was done on ice. A single cell suspension was derived by adding 5ml TryPle (1×) and incubation at 37 °C for 10 minutes. Cells were then transferred to a 15 ml tube and were centrifuged at 1200 for 5 minutes and 20 °C. The pellet was re-suspended in 90 µL FACS buffer plus 10 µL of FcR-blocking reagent (Miltenyi Biotec) and incubated at 4 °C for 10 minutes. Then, cells were centrifuged in 1200 RCF for 5 minutes and then resuspended in 200 µL of FACS buffer and were transferred on ice to sort on the FACS machine. Cells were sorted directly into the Trizol.
Magnetic Cell Sorting (MACS)
The MACS CD34 MicroBead Kit (Miltenyi Biotec) was used to purify the CD34-expressing cells. After making a single suspension similar to FACS, the cell then was filtered by Pre-separation filters, 30 µm (Miltenyi Biotec) to prevent cell clumps from clogging the magnetic column, then centrifuged and re-suspended in MACS buffer and counted. Then 100ul MicroBeads conjugated to monoclonal mouse anti-human CD34 antibodies (isotype: mouse IgG1) were added and were incubated at 4 °C for 30 minutes. Then cells were centrifuged and re-suspended in MACS buffer and proceeded for magnetic separation with MS columns (Miltenyi Biotec). Cells conjugated with CD34 MicroBeads were loaded into the reservoir of the column. The unlabeled cells flow through the column collected in a 15ml centrifuge tube. Finally, The MS magnetic column was removed from the magnetic field, 500 ul of MACS buffer was loaded in the column reservoir, and magnetically labeled cells were flushed out of the column by pushing the plunger steadfastly into the reservoir of the column.
Extraction of RNA for CD34+ and CD34- cells
During FACS, cells were directly sorted into 1 ml of Trizol (Sigma-Aldrich) and then were transferred into 1.5 ml Eppendorf and rotated in a shaker for 10 minutes in RT to allow lysis to start. Then 200 µL Chloroform (VWR, UK) was added to each Eppendorf, mixed completely, and left at RT for 10 minutes. Tubes were then centrifuged at 12000 RCF for 15 minutes at 4 °C. The top aqueous phase including RNA was removed and placed into the new tube. 1ul of RNase-free glycogen (Ambion, UK) and 500 µL of Isopropanol (VWR, UK) were added to precipitate the RNA. Tubes were inverted to mix and incubated at RT for 10 minutes and then were centrifuged at 12000 RCF at 4 °C for 10 minutes. After the centrifuge, the supernatant was poured off and 1ml of 70% Ethanol was added, completely mixed with vortex, and centrifuged at 4 °C at 7400 RCF for 5 minutes. after the last centrifuge, the supernatant was poured off and the pellet was left to airdry. Then pellet was re-suspended in 21µL of RNAssecure (Ambion, UK) and was heated to 50 °C for 10 minutes.
RNA-sequencing
In our project, Illumina TruSeq RNA v2 (Wang, et al. 2009) RNA sequencing protocol was used. As a starting material, approximately 250 ng of Trizol-extracted RNA was sent to the sequencing facility of the UCL Institute of Child Health. Before RNA sequencing, samples were tested for quality using Agilent RNA Integrity Number (RIN) to determine RNA quality for RNA sequencing analysis. After quality control, RNA samples were further proceeded for library preparation and sequencing on the Illumina TruSeq RNA v2 platform.
The project described was supported by the UCL Institute of Ophthalmology and the National Centre for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR002002. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Core Grant for Vision Research EY01792 (MIR) from NEI/NIH; Research to Prevent Blindness Unrestricted Departmental Grant.
Author contributions
E.J. R.M.M. and W. R conducted the experiments. W.R. and E.J. analyzed the data. E.J. wrote and supervised the manuscript.
- Hassanpour M, Salybekov AA, Kobayashi S, Asahara T. CD34 positive cells as endothelial progenitor cells in biology and medicine. Front Cell Dev Biol. 2023 Apr 17;11:1128134. doi: 10.3389/fcell.2023.1128134. PMID: 37138792; PMCID: PMC10150654.
- Leeper NJ, Hunter AL, Cooke JP. Stem cell therapy for vascular regeneration: adult, embryonic, and induced pluripotent stem cells. Circulation. 2010 Aug 3;122(5):517-26. doi: 10.1161/CIRCULATIONAHA.109.881441. PMID: 20679581; PMCID: PMC2920605.
- Xie Y, Fan Y, Xu Q. Vascular Regeneration by Stem/Progenitor Cells. Arterioscler Thromb Vasc Biol. 2016 May;36(5):e33-40. doi: 10.1161/ATVBAHA.116.307303. PMID: 27122628.
- Hong X, Le Bras A, Margariti A, Xu Q. Reprogramming towards endothelial cells for vascular regeneration. Genes Dis. 2016 Feb 27;3(3):186-197. doi: 10.1016/j.gendis.2016.02.003. PMID: 30258888; PMCID: PMC6147164.
- Masuda H, Iwasaki H, Kawamoto A, Akimaru H, Ishikawa M, Ii M, Shizuno T, Sato A, Ito R, Horii M, Ishida H, Kato S, Asahara T. Development of serum-free quality and quantity control culture of colony-forming endothelial progenitor cell for vasculogenesis. Stem Cells Transl Med. 2012 Feb;1(2):160-71. doi: 10.5966/sctm.2011-0023. Epub 2012 Feb 6. PMID: 23197763; PMCID: PMC3659679.
- Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997 Feb 14;275(5302):964-7. doi: 10.1126/science.275.5302.964. PMID: 9020076.
- Otani A, Kinder K, Ewalt K, Otero FJ, Schimmel P, Friedlander M. Bone marrow-derived stem cells target retinal astrocytes and can promote or inhibit retinal angiogenesis. Nat Med. 2002 Sep;8(9):1004-10. doi: 10.1038/nm744. Epub 2002 Jul 29. PMID: 12145646.
- Otani A, Dorrell MI, Kinder K, Moreno SK, Nusinowitz S, Banin E, Heckenlively J, Friedlander M. Rescue of retinal degeneration by intravitreally injected adult bone marrow-derived lineage-negative hematopoietic stem cells. J Clin Invest. 2004 Sep;114(6):765-74. doi: 10.1172/JCI21686. PMID: 15372100; PMCID: PMC516263.
- Cho HJ, Lee N, Lee JY, Choi YJ, Ii M, Wecker A, Jeong JO, Curry C, Qin G, Yoon YS. Role of host tissues for sustained humoral effects after endothelial progenitor cell transplantation into the ischemic heart. J Exp Med. 2007 Dec 24;204(13):3257-69. doi: 10.1084/jem.20070166. Epub 2007 Dec 10. PMID: 18070934; PMCID: PMC2150988.
- Fadini GP. Circulating stem cells and cardiovascular outcomes: from basic science to the clinic. Eur Heart J. 2019.
- Rennert RC, Sorkin M, Garg RK, Gurtner GC. Stem cell recruitment after injury: lessons for regenerative medicine. Regen Med. 2012 Nov;7(6):833-50. doi: 10.2217/rme.12.82. PMID: 23164083; PMCID: PMC3568672.
- Strilić B, Kucera T, Eglinger J, Hughes MR, McNagny KM, Tsukita S, Dejana E, Ferrara N, Lammert E. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev Cell. 2009 Oct;17(4):505-15. doi: 10.1016/j.devcel.2009.08.011. PMID: 19853564.
- Nielsen JS, McNagny KM. Novel functions of the CD34 family. J Cell Sci. 2008 Nov 15;121(Pt 22):3683-92. doi: 10.1242/jcs.037507. Erratum in: J Cell Sci. 2008 Dec 15;121(Pt 24):4145. PMID: 18987355.
- Salybekov AA, Kobayashi S, Asahara T. Characterization of Endothelial Progenitor Cell: Past, Present, and Future. Int J Mol Sci. 2022 Jul 12;23(14):7697. doi: 10.3390/ijms23147697. PMID: 35887039; PMCID: PMC9318195.
- Kawamoto A, Katayama M, Handa N, Kinoshita M, Takano H, Horii M, Sadamoto K, Yokoyama A, Yamanaka T, Onodera R, Kuroda A, Baba R, Kaneko Y, Tsukie T, Kurimoto Y, Okada Y, Kihara Y, Morioka S, Fukushima M, Asahara T. Intramuscular transplantation of G-CSF-mobilized CD34(+) cells in patients with critical limb ischemia: a phase I/IIa, multicenter, single-blinded, dose-escalation clinical trial. Stem Cells. 2009 Nov;27(11):2857-64. doi: 10.1002/stem.207. PMID: 19711453.
- Tanaka R, Masuda H, Kato S, Imagawa K, Kanabuchi K, Nakashioya C, Yoshiba F, Fukui T, Ito R, Kobori M, Wada M, Asahara T, Miyasaka M. Autologous G-CSF-mobilized peripheral blood CD34+ cell therapy for diabetic patients with chronic nonhealing ulcer. Cell Transplant. 2014 Feb;23(2):167-79. doi: 10.3727/096368912X658007. Epub 2012 Oct 25. PMID: 23107450.
- Lee JH, Lee SH, Yoo SY, Asahara T, Kwon SM. CD34 hybrid cells promote endothelial colony-forming cell bioactivity and therapeutic potential for ischemic diseases. Arterioscler Thromb Vasc Biol. 2013 Jul;33(7):1622-34. doi: 10.1161/ATVBAHA.112.301052. Epub 2013 May 2. PMID: 23640491.
- Aries A, Vignon C, Zanetti C, Goubaud A, Cormier A, Diederichs A, Lahlil R, Hénon P, Garitaonandia I. Development of a potency assay for CD34+ cell-based therapy. Sci Rep. 2023 Nov 11;13(1):19665. doi: 10.1038/s41598-023-47079-8. PMID: 37952030; PMCID: PMC10640600.
- AbuSamra DB, Aleisa FA, Al-Amoodi AS, Jalal Ahmed HM, Chin CJ, Abuelela AF, Bergam P, Sougrat R, Merzaban JS. Not just a marker: CD34 on human hematopoietic stem/progenitor cells dominates vascular selectin binding along with CD44. Blood Adv. 2017 Dec 26;1(27):2799-2816. doi: 10.1182/bloodadvances.2017004317. PMID: 29296932; PMCID: PMC5745127.
- Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 2014 Jun;32(6):1380-9. doi: 10.1002/stem.1661. PMID: 24497003; PMCID: PMC4260088.
- Hughes MR, Canals Hernaez D, Cait J, Refaeli I, Lo BC, Roskelley CD, McNagny KM. A sticky wicket: Defining molecular functions for CD34 in hematopoietic cells. Exp Hematol. 2020 Jun;86:1-14. doi: 10.1016/j.exphem.2020.05.004. Epub 2020 May 16. PMID: 32422232.
- Anjos-Afonso F, Bonnet D. Human CD34+ hematopoietic stem cell hierarchy: how far are we with its delineation at the most primitive level? Blood. 2023 Aug 10;142(6):509-518. doi: 10.1182/blood.2022018071. PMID: 37018661; PMCID: PMC10644061.
- Kawamoto A, Gwon HC, Iwaguro H, Yamaguchi JI, Uchida S, Masuda H, Silver M, Ma H, Kearney M, Isner JM, Asahara T. Therapeutic potential of ex vivo expanded endothelial progenitor cells for myocardial ischemia. Circulation. 2001 Feb 6;103(5):634-7. doi: 10.1161/01.cir.103.5.634. PMID: 11156872.
- Kawamoto A, Asahara T, Losordo DW. Transplantation of endothelial progenitor cells for therapeutic neovascularization. Cardiovasc Radiat Med. 2002 Jul-Dec;3(3-4):221-5. doi: 10.1016/s1522-1865(03)00082-9. PMID: 12974378.
- Yang J, Ii M, Kamei N, Alev C, Kwon SM, Kawamoto A, Akimaru H, Masuda H, Sawa Y, Asahara T. CD34+ cells represent highly functional endothelial progenitor cells in murine bone marrow. PLoS One. 2011;6(5):e20219. doi: 10.1371/journal.pone.0020219. Epub 2011 May 31. PMID: 21655289; PMCID: PMC3105013.
- Rahat MA, Hemmerlein B, Iragavarapu-Charyulu V. The regulation of angiogenesis by tissue cell-macrophage interactions. Front Physiol. 2014 Jul 9;5:262. doi: 10.3389/fphys.2014.00262. PMID: 25071604; PMCID: PMC4087329.
- Corliss BA, Azimi MS, Munson JM, Peirce SM, Murfee WL. Macrophages: An Inflammatory Link Between Angiogenesis and Lymphangiogenesis. Microcirculation. 2016 Feb;23(2):95-121. doi: 10.1111/micc.12259. PMID: 26614117; PMCID: PMC4744134.
- Ramirez-Pedraza M, Fernández M. Interplay Between Macrophages and Angiogenesis: A Double-Edged Sword in Liver Disease. Front Immunol. 2019 Dec 12;10:2882. doi: 10.3389/fimmu.2019.02882. PMID: 31921146; PMCID: PMC6927291.
- Jiang L, Chen T, Sun S, Wang R, Deng J, Lyu L, Wu H, Yang M, Pu X, Du L, Chen Q, Hu Y, Hu X, Zhou Y, Xu Q, Zhang L. Nonbone Marrow CD34+ Cells Are Crucial for Endothelial Repair of Injured Artery. Circ Res. 2021 Oct;129(8):e146-e165. doi: 10.1161/CIRCRESAHA.121.319494. Epub 2021 Sep 3. PMID: 34474592.
- Lu SJ, Feng Q, Caballero S, Chen Y, Moore MA, Grant MB, Lanza R. Generation of functional hemangioblasts from human embryonic stem cells. Nat Methods. 2007 Jun;4(6):501-9. doi: 10.1038/nmeth1041. Epub 2007 May 7. PMID: 17486087; PMCID: PMC3766360.
- Glaser DE, Gower RM, Lauer NE, Tam K, Blancas AA, Shih AJ, Simon SI, McCloskey KE. Functional characterization of embryonic stem cell-derived endothelial cells. J Vasc Res. 2011;48(5):415-28. doi: 10.1159/000324752. Epub 2011 May 31. PMID: 21625175; PMCID: PMC3121553.
- Timmermans F, Plum J, Yöder MC, Ingram DA, Vandekerckhove B, Case J. Endothelial progenitor cells: identity defined? J Cell Mol Med. 2009 Jan;13(1):87-102. doi: 10.1111/j.1582-4934.2008.00598.x. PMID: 19067770; PMCID: PMC3823038.
- Orlova VV, Drabsch Y, Freund C, Petrus-Reurer S, van den Hil FE, Muenthaisong S, Dijke PT, Mummery CL. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol. 2014 Jan;34(1):177-86. doi: 10.1161/ATVBAHA.113.302598. Epub 2013 Oct 24. PMID: 24158517.
- Prasain N, Lee MR, Vemula S, Meador JL, Yoshimoto M, Ferkowicz MJ, Fett A, Gupta M, Rapp BM, Saadatzadeh MR, Ginsberg M, Elemento O, Lee Y, Voytik-Harbin SL, Chung HM, Hong KS, Reid E, O'Neill CL, Medina RJ, Stitt AW, Murphy MP, Rafii S, Broxmeyer HE, Yoder MC. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat Biotechnol. 2014 Nov;32(11):1151-1157. doi: 10.1038/nbt.3048. Epub 2014 Oct 12. PMID: 25306246; PMCID: PMC4318247.
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16806-11. doi: 10.1073/pnas.0603916103. Epub 2006 Oct 31. PMID: 17077151; PMCID: PMC1636536.
- Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008 Jan 10;2(1):60-71. doi: 10.1016/j.stem.2007.10.011. PMID: 18371422; PMCID: PMC2533280.
- Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, Ding M, Deng H. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008 Feb 15;111(4):1933-41. doi: 10.1182/blood-2007-02-074120. Epub 2007 Nov 27. PMID: 18042803.
- Bai H, Xie YL, Gao YX, Cheng T, Wang ZZ. The balance of positive and negative effects of TGF-β signaling regulates the development of hematopoietic and endothelial progenitors in human pluripotent stem cells. Stem Cells Dev. 2013 Oct 15;22(20):2765-76. doi: 10.1089/scd.2013.0008. Epub 2013 Jul 19. PMID: 23758278; PMCID: PMC3787486.
- Mayer H, Bertram H, Lindenmaier W, Korff T, Weber H, Weich H. Vascular endothelial growth factor (VEGF-A) expression in human mesenchymal stem cells: autocrine and paracrine role on osteoblastic and endothelial differentiation. J Cell Biochem. 2005 Jul 1;95(4):827-39. doi: 10.1002/jcb.20462. PMID: 15838884.
- Holmes DI, Zachary I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. 2005;6(2):209. doi: 10.1186/gb-2005-6-2-209. Epub 2005 Feb 1. PMID: 15693956; PMCID: PMC551528.
- Bautch VL. VEGF-directed blood vessel patterning: from cells to organism. Cold Spring Harb Perspect Med. 2012 Sep 1;2(9):a006452. doi: 10.1101/cshperspect.a006452. PMID: 22951440; PMCID: PMC3426816.
- Balaji S, King A, Crombleholme TM, Keswani SG. The Role of Endothelial Progenitor Cells in Postnatal Vasculogenesis: Implications for Therapeutic Neovascularization and Wound Healing. Adv Wound Care (New Rochelle). 2013 Jul;2(6):283-295. doi: 10.1089/wound.2012.0398. PMID: 24527350; PMCID: PMC3751266.
- Rehman J, Li J, Orschell CM, March KL. Peripheral blood "endothelial progenitor cells" are derived from monocyte/macrophages and secrete angiogenic growth factors. Circulation. 2003 Mar 4;107(8):1164-9. doi: 10.1161/01.cir.0000058702.69484.a0. PMID: 12615796.
- van den Berg BH, Thanthiriwatte C, Manda P, Bridges SM. Comparing gene annotation enrichment tools for functional modeling of agricultural microarray data. BMC Bioinformatics. 2009 Oct 8;10 Suppl 11(Suppl 11):S9. doi: 10.1186/1471-2105-10-S11-S9. PMID: 19811693; PMCID: PMC3226198.
- Newman AM, Cooper JB. AutoSOME: a clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinformatics. 2010 Mar 4;11:117. doi: 10.1186/1471-2105-11-117. PMID: 20202218; PMCID: PMC2846907.
- Ringnér M. What is principal component analysis? Nat Biotechnol. 2008 Mar;26(3):303-4. doi: 10.1038/nbt0308-303. PMID: 18327243.
- Park KS. Tgf-Beta family signaling in embryonic stem cells. Int J Stem Cells. 2011 Jun;4(1):18-23. doi: 10.15283/ijsc.2011.4.1.18. PMID: 24298330; PMCID: PMC3840975.
- Pardali E, Goumans MJ, ten Dijke P. Signaling by members of the TGF-beta family in vascular morphogenesis and disease. Trends Cell Biol. 2010 Sep;20(9):556-67. doi: 10.1016/j.tcb.2010.06.006. Epub 2010 Jul 23. PMID: 20656490.
- Kimelman D. Mesoderm induction: from caps to chips. Nat Rev Genet. 2006 May;7(5):360-72. doi: 10.1038/nrg1837. PMID: 16619051.
- Omelyanchuk N, Orlovskaya IA, Schraufstatter IU, Khaldoyanidi SK. Key players in the gene networks guiding ESCs toward mesoderm. J Stem Cells. 2009;4(3):147-60. PMID: 20232600; PMCID: PMC2888472.
- Johnson KE, Wilgus TA. Vascular Endothelial Growth Factor and Angiogenesis in the Regulation of Cutaneous Wound Repair. Adv Wound Care (New Rochelle). 2014 Oct 1;3(10):647-661. doi: 10.1089/wound.2013.0517. PMID: 25302139; PMCID: PMC4183920.
- Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med. 2012 Jul;2(7):a006502. doi: 10.1101/cshperspect.a006502. PMID: 22762016; PMCID: PMC3385940.
- Thippabhotla S, Zhong C, He M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep. 2019 Sep 10;9(1):13012. doi: 10.1038/s41598-019-49671-3. PMID: 31506601; PMCID: PMC6736862.
- Vila E, Solé M, Masip N, Puertas-Umbert L, Amaro S, Dantas AP, Unzeta M, D'Ocon P, Planas AM, Chamorro Á, Jiménez-Altayó F. Uric acid treatment after stroke modulates the Krüppel-like factor 2-VEGF-A axis to protect brain endothelial cell functions: Impact of hypertension. Biochem Pharmacol. 2019 Jun;164:115-128. doi: 10.1016/j.bcp.2019.04.002. Epub 2019 Apr 4. PMID: 30954486.
- Medina RJ, O'Neill CL, Sweeney M, Guduric-Fuchs J, Gardiner TA, Simpson DA, Stitt AW. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med Genomics. 2010 May 13;3:18. doi: 10.1186/1755-8794-3-18. PMID: 20465783; PMCID: PMC2881111.
- Dunne MR, Michielsen AJ, O'Sullivan KE, Cathcart MC, Feighery R, Doyle B, Watson JA, O'Farrell NJ, Ravi N, Kay E, Reynolds JV, Ryan EJ, O'Sullivan J. HLA-DR expression in tumor epithelium is an independent prognostic indicator in esophageal adenocarcinoma patients. Cancer Immunol Immunother. 2017 Jul;66(7):841-850. doi: 10.1007/s00262-017-1983-1. Epub 2017 Mar 18. PMID: 28315927; PMCID: PMC5489642.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
