Open Journal of Biological Sciences
Identification of genetic differences between two species of morphologically identical Metapenaeus Genus from different environmental locations (Khaur Abdullah & Shatt Al- Arab) Southern of Iraq using (RFLP & RAPD) PCR molecular markers
Rabeeha Alzuhairi*
Cite this as
Alzuhairi R (2020) Identification of genetic differences between two species of morphologically identical Metapenaeus Genus from different environmental locations (Khaur Abdullah & Shatt Al- Arab) Southern of Iraq using (RFLP & RAPD) PCR molecular markers. Open J Biol Sci 5(1): 015-021. DOI: 10.17352/ojbs.000018Metapenaeus affinis a species from Metapenaeus genera of Penaeidae shrimps in khaur Abdullah (salty water southern of Iraq), morphologically closely related organism with suspected member in Shatt al-Arab(freshwater)also southern of Iraq. Genetic Characterization for them was study to determine the genetically differences by using (RFLP & RAPD)/PCR molecular markers. The genetic heterogeneity at the DNA level was high in the two organisms in spite of Metapenaes genus share considerable external features. In RFLP marker, voucher DAAPV F7 in 16S rRNA gene was used which was amplified then digested by using the enzymes (SmI ,TagI and HindIII). Restriction patterns of TaqI, SmI & Hind III digested voucher DAAPV F7 in 16S rRNA gene, were clearly differentiate both Metapenaeus affinis and suspected one. Size pieces which were cut result in differing size and differing numbers of patterns Whereas a species which suspected to be Metapenaeus affinis morphologically, gave different results. Overlapping patterns between different species not observed. RFLP results were confirmed the RAPD-PCR results where PCR was carried out using standers conditions for 20 random oligonucleotide primers on gDNA samples of two organisms .Out of them, twelve primers produced high reproducibility and consistent with RAPD profile and yielded monomorphic as well as polymorphic fragments ,while 8 primers yielded either weak amplifications with ambiguity or no amplification at both. The present RAPD study had shown a higher level of polymorphism in the two members of the Metapenaeus genus. 12 random primers yielded a total of 82 fragments, of which, 50 bands were polymorphic whereas 32 fragments were monomorphic. Both molecular markers (RAPD and RFLP/PCR ) were facilitated the task of distinguishing species that are morphologically close species and very useful in determining the distribution of the exotic species.
Introduction
Shatt al-Arab (River of the Arabs) is a river in Southwest Asia of some 200 km (120 mi) in length, formed by the confluence of the Euphrates and the Tigris in the town of al-Qurnah in the Basra Governorate of southern Iraq. The southern end of the river constitutes the border between Iraq and Iran down to the mouth of the river as it discharges into the Arabian Gulf. It varies in width from about 232 metres (761 ft) at Basra to 800 metres (2,600 ft) at its mouth. It is thought that the waterway formed relatively recently in geologic time, with the Tigris and Euphrates originally emptying into the Arabian Gulf via a channel further to the west.
Indeed, the marshlands in Iraq were drained in the early 1990s in order to increase government control over the Arab Shiites (Marsh Arabs) who lived there. Restoration of the marshlands began in 2003, following the invasion of Iraq by Anglo-American forces, but only half the area has been restored. The river supplies fresh water to Iraq and Kuwait but the construction of dams and the demand for water upstream has led to a greatly increased salt content. The Shatt al Arab is navigable for oceangoing vessels as far as Basra, Iraq’s chief port.
In Iraqi south waters there are two main areas are considered important regions for study and research ongoing because of environmental and life changes that occur from time to time for several reasons, which are (Khour Abdullah and Shatt al-Arab) which is always the focus of attention of scientists and researchers, particularly in the south of Iraq there are aquatic organisms variety. Aquatic organisms living in salty water environment and aquatic organisms live in fresh water environment and the other can live in salty and fresh water environment which are called (euryhaline). Often most scientists suspect of phenotypic classification of some aquatic organisms and which are too closed, scientist classified it according to potentially classification. Through present study resorted to solve this problem genetically.
In many countries, Metapenaeus genus classified into several species, some countries relied on phenotypic classification and other countries adopted genetically classification but other countries relied on both. Iraq has been registered only one species of Metapeaeus genus (Metapenaeus affinis ) which lives in salty water, (Khour Abd Allah or Al-faw) but the puzzle in species lives in fresh water (Shatt al-Arab, or marsh) which is closed Metapenaeus affinis species in number of phenotypic traits.
The diversity of organisms is influenced by multiple evolutionary factors, a situation that can affect the morphology, ecosystems and other biological behaviors of a plant or animal. Biological diversity is evident in a clear majority of species and leads to individual variations in numerous characteristics [1]. Differences in species because of diversity is reflected in the genetic differences and environmental factors, or a combination of both. Metapenaeus is a genus of pawns and has been classified into different species including the Metapenaeus affinis, a marine water pawn [2]. While the marine pawn has been defined and named, a second closely related organism has been discovered in freshwater bodies. Unlike the marine pawn, it is small but has similar morphological characteristics like the marine counterpart [3]. To compare the genetic diversity, different molecular biology techniques, apart from the current conventional genetic analyzer capillary sequencer can be used [4]. Understanding the genetic diversity of the two organisms is critical in categorizing them and creating a new taxonomic characterization of the freshwater species.
Molecular biology techniques in genetic diversity of Metapenaeus species
Molecular characterization of different species is gene dependent, an approach that focuses on the differences in DNA code at different loci. Metapenaeus species may have arisen from the same organism but experienced environmental pressures, leading to mutations which may have enabled the unknown species to be smaller and to survive in freshwater bodies. Gene mutation or chromosomal changes are common contributors of genetic diversity and are associated with biological events such as meiosis and fertilization. To understand the molecular biodiversity of the two organisms, the differences in DNA base sequence or the amino acid of different protein can be assessed. Different techniques have so far been developed that can be used for the molecular characterization of diversity between the two species. While DNA sequencing is the most commonly used technique today, another cost-effective and non-laborious techniques also exist. Rapid Amplified Polymorphic DNA (RAPD) technique is one method that can be adopted in the diversity characterization of the organisms [5]. RAPD data used and supported by RFLP-PCR marker which used 16s rRNA gene. In this method, genomic DNAwas amplified using specific primers to identify possible polymorphic markers in the absence of prior information on the mitochondrial genetic loci, according to Alex and Kochzius [6].
To define the genetic difference between the two organisms of shrimp using RAPD, many steps were followed. First, DNA was extracted from the two organisms according to standard procedure [7]. The process was avoiding potential breakdown of the DNA which can affect the amplification and identification of polymorphic sites [8]. Once DNA extracted, the next step involves the introduction of single arbitrary primers. Arbitrary primer amplification is a technology that has traditionally been used in the classification of Bacillus strains when DNA extraction is done from bacterial colonies with single templates [9]. Primers are developed arbitrarily without the specific target of loci. Knowledge of the genetic composition of Metapenaeus marine species is used in the development of primers [10]. Furthermore, the primers specified to target specific conserved sequences to define homology and demonstrate an area of evolutionary diversion.
Once primers introduced, the next step was the amplification through polymerase chain reaction (PCR). The amplification process duplicates the genetic loci that bind the primers, further increasing the number of copies. Once the fragments were amplified, separation for the different samples was undertaken using gel electrophoresis, a process that exploits the charge and size of the fragments [11]. A kilo base lambda DNA was used in the gel to help in the determination of the fragment sizes [10]. Once the fragments separated on 1% agarose gel in the presence of ethidium bromide, visualization was done under UV light. The band that moves highest has the least size and comparison was done for the two organisms [3,12]. Size fragments were compared in the gel to determine the presence of microsatellites and polymorphic sites which act the molecular markers. The use of RAPD identified potential polymorphic markers that can differentiate the two Metapenaeus organisms and help demonstrate the difference in ecosystems. The method was convenient for this characterization due to its quick, simple and efficient nature. The process was only dependent on the thermocycling machine and the agarose gel electrophoresis equipment [13]. However, prior knowledge on the size of conserved sequences may be needed to compare and identify points of polymorphic diversion [14].
Materials and methods
Species collection and identification
A total of thirty-five specimens of Metapenaeus affinis species as well as the same number of suspection organism were collected from two different locations, Khour Abd Allah and Shatt al-Arab which situated in Basra southern of Iraq. Metapenaeus affinis situated in Khour Abd Allah water (salty water) whereas the other suspected shrimp is situated in Shatt al-Arab (freshwater) as shown in a Figure 1.
Genomic DNA isolation
Total genomic DNA was isolated from a piece of pleopod of each samples of shrimp using a phenol-chloroform-proteinase K method [15], DNeasy Tissue Kit (Germany). DNA concentrations were spectrophotometrically determined at using a NanoDrop; 1000 Spectrophotometer at the absorbance of 260 (A260) and 280 nm (A280). The purity of extracted DNA was determined by using A260/A280 ratio. 1% Agarose Gel Electrophoresis was performed to detect the genomic DNA using Gel documentation. The DNA was diluted using TE buffer to a final concentration of µg/ml for RAPD analysis. It was then supported by using the PCR-RFLP technique. Twelve random oligonucleotide primers were used for RAPD-PCR analysis and specific primers were used to amplified voucher DAAPV F7 in 16s rRNA gene for PCR-RFLP marker. The sequences of these primers for RAPD technique was given in Table 1. PCR reactions of RAPD marker were performed in 25 µl reaction volumes containing 1x PCR buffer (100 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2 and 50 mM , KCl, 0.2 µM primer,100 µm each of dATP, dCTP, dGTP and dTTP 0.75 U of Taq DNA polymerase 20 ng of template DNA [7]. Thermo cycler conditions were as follows; for initial denaturation at 94ºC for 3 min, followed by 35ºC cycles at 94ºC for 1 min, 36ºC for 1 min and 72ºC for 2 min, one cycle at 72ºC for 10 min, and then 4ºC soak. The amplified samples were stored at-20ºC for further analysis. RAPD reaction products were visualized on 1.5% agarose gel, amplified fragments were identified by its size in base pairs and its associated primer. For each primer, PCR amplified products were scored on the gel images for monomorphic or polymorphic bands of a given amplification product of each sample of two organisms. Then RAPD-PCR analysis was supported by PCR-RFLP marker which used voucher DAAPV F7 in16S rRNA gene. The 16s rRNA gene fragment here seems suitable for examining phylogenetic relationships at the species or genus levels in Crustaceans. The 16s rDNA sequence has hypervariable regions, where sequences have diverged over evolutionary time. Strongly conserved regions often flank these hypervariable regions. (PCR)was used to amplify the voucher DAAPV F7 in 16s rRNA gene, in order to search for molecular markers that could discriminate both species. The primers were designed based on rRNA gene sequences found in GenBank where nucleotide sequences of the voucher DAAPV F7 in 16S rRNA gene were aligned using Clustal W [16]. A pair of primers primed at the conserved regions of 16S rRNA was designed and tested against 70 shrimp individuals. Namely as following:
OLIGO start len tm gc% any 3’ seq
LEFT PRIMER 67 20 60.12 50.00 3.00 3.00 CCGTGCGAAGGTAGCATAAT
RIGHT PRIMER 302 20 59.92 50.00 4.00 1.00 TATATTCTCGTCGCCCCAAC
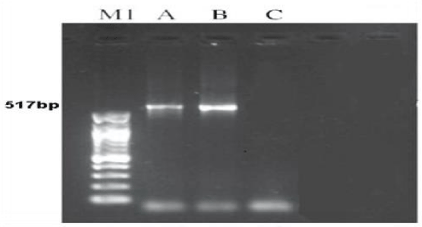
The target DNA fragments (voucher DAAPV F7) for both organisms were amplified a 517bp segment of the 16s rRNA gene. PCR reaction was carried out in Eppendorf Thermo Cycler. [17]. The reactions in volumes of 50μL containing 4μl of DNA template, 4μl of each primers, 25μl of master mix completed the size with 18μl of dd.w. Thermo cycler conditions were as follows: 5min at 95°C for pre-running, then 35cycles at 95°C for denaturation, 40s at 50-52 °C for annealing, and 40s at 72 °C for extension followed by 10 min at 72 °C for a final extension. Amplicons were visualized on 1.2% agarose Gel as shown in Figure 14. Then voucher DAAPV F7 in 16s rRNA gene amplification products were digested with TaqI (TCGA), Sm II (TCYRAG) and Hind III (AAGCTT). The restricted products were electrophoresed through 2.0% agarose gel and visualized under a UV transilluminator after ethidium bromide staining [18].
Results and discussion
PCR was carried out using standers conditions for twenty random oligonucleotide primers on gDNA samples of two organisms (Metapenaeus affinis & suspected shrimp). Out of twenty primers, twelve primers produced high reproducibility and consistent with RAPD profile and yielded monomorphic as well as polymorphic fragments, while 8 primers yielded either weak amplifications with ambiguity or no amplification at all. The present RAPD study had shown a higher level of polymorphism in the two members of the Penaeidae. Twelve random primers yielded a total of 82 fragments, of which, 50 bands were polymorphic while 32 bands were monomorphic were shared by both species of the organisms as given in Tables 1,2.
The number and size of the amplified products varied depending on genetic characterization of the species. A unique band of 1892 bp obtained for Metapenaeus affinius which was highly species specific for primer OPF-09 and not both organisms shared in this primer. Maximum of 11 bands obtained from OPA-13 primer and the least of three bands from OPC-02 primer. Highly polymorphic bands obtained with the primer OPA-13. When the size of the fragment is concerned, the highest range of 164-1892 obtained from the OPF-09 whereas the lowest range from OPE-02 (269-1207). RAPD profiles obtained by twelve primers were shown in the Figures 1-13 . Maximum of 11 bands obtained from OPA-13 primer and the least of 3 bands from OPC-02 primer. Species-diagnostic markers from DNA segments should exhibiting low genetic polymorphism within a particular species but showing high genetic divergence between different species [15].
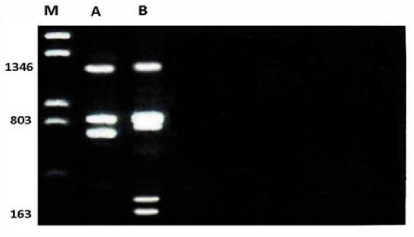
Profile of RAPD obtained by OPA-08 primer as shown in the Figure 2. The size of the amplified products ranged from 163-1346bp. The smallest fragment belongs to Unknown member and the biggest one belongs to both members. A total of 6 bands were scored. Of these bands, two were monomorphic with 803 and 1346bp and shared both members (species). Out of 7 bands, 4 bands were polymorphic and shared by both species.
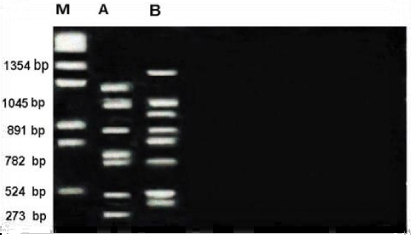
RAPD profile obtained by OPA-13 primer as shown in the Figure 3. The size of the amplified products ranged from 273-1354bp. The smallest fragment belongs to Metapenaeus affinis and the biggest one belongs to unknown member. A considerable amount of polymorphism was detected with this primer. A total of 11 bands were scored. Of these bands, four were monomorphic with 524,782,891 and 1045bp and shared by both species. Out of 11 bands, 7 bands were polymorphic and shared by both species.
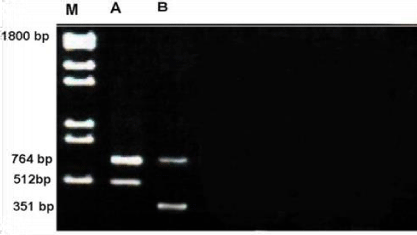
Profile of RAPD obtained by OPC-02 primer is depicted in the Figure 4. The size of the amplified products ranged from 351-1800 bp. The smallest fragment belongs to unknown member whereas both members had the largest fragment.
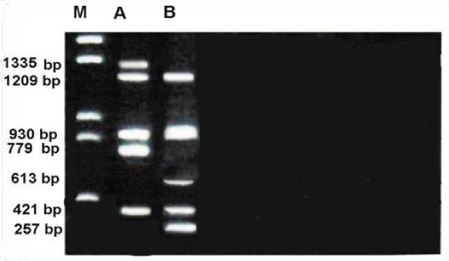
Figure 5 shows RAPD profile obtained by OPD-08 primer. The size of the amplified products ranged from 57-1335bp. The smallest fragment was observed for unknown member and the largest to Metapenaeus affinis. Four of polymorphism was detected with this primer. And 3 of monomorphic of 7 total fragments were scored.
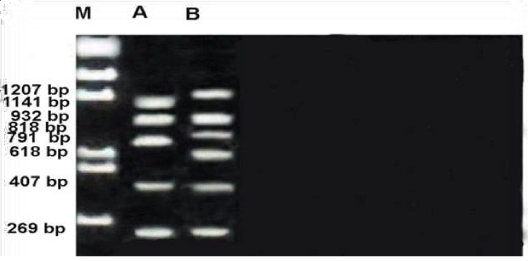
RAPD profile of OPE-02 primer is depicted in the Figure 6. The size of the amplified products ranged from 269-1207bp. unknown member with the largest band and both members with smallest monomorphic.A total of 8 fragments were scored. Of these fragments, three were monomorphic band with 269,407 and 932bp shared by both species.
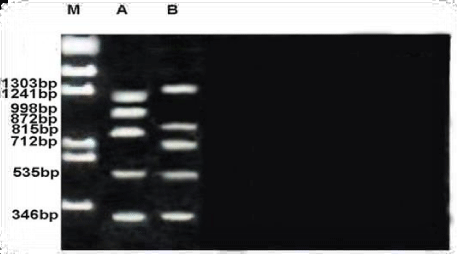
The size of the amplified products of RAPD profile obtained by OPE-03 primer ranged from 346-1303bp. Both members had small fragment whereas the biggest one observed in unknown member. A considerable amount of polymorphism was detected with this primer. Of the eight fragments scored, two were monomorphic (346 &535bp) shared by both species and six fragments were polymorphic and shared by both species as shown in the Figure 7.
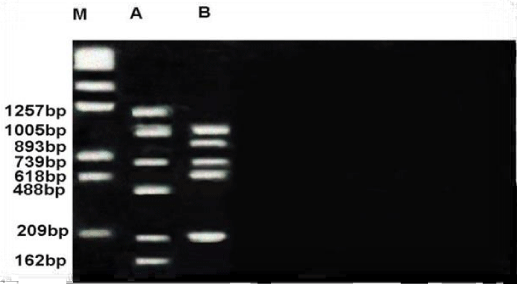
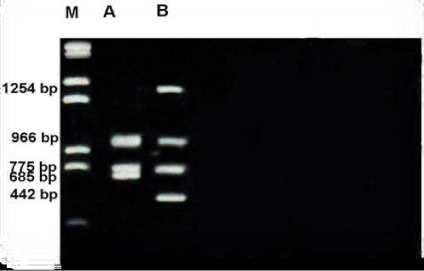
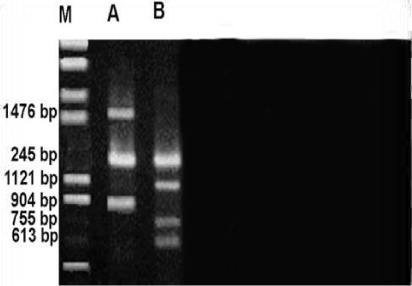
Figure 8 shows RAPD profile obtained by OPE-06 primer. The size of the amplified products ranged from 162-1257bp. Metapenaeus affinis had the smallest fragment and it also had the biggest fragment. A total of 8 fragments were scored, of these fragments, three were monomorphic with 209,739 &1005bp shared by both species. five fragments were polymorphic and shared by both species. RAPD profile obtained by OPF-05 primer was presented in the Figure 9. The size of the amplified products ranged from 442-1254bp. Unknown member had smallest fragment as well as it had the biggest one. A total of 5 fragments were scored of these fragments, two were monomorphic band with 775 and 966 bp shared by both species. Out of 5 fragments, three were polymorphic, shared by both species. Figure 10 shows RAPD profile obtained by OPF-06 primer. The size of the amplified products ranged from 284-1476bp. Unknown member had the smallest fragment and Metapenaeus affinis species had the biggest fragment. A total of 7 fragments were scored, of these fragments, one was monomorphic with 1245bp shared by both species. Six fragments were polymorphic and shared by both species.
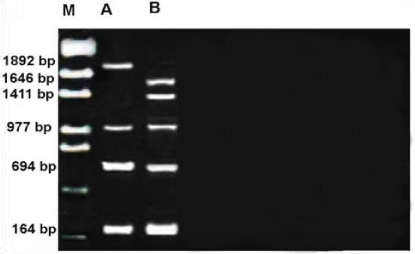
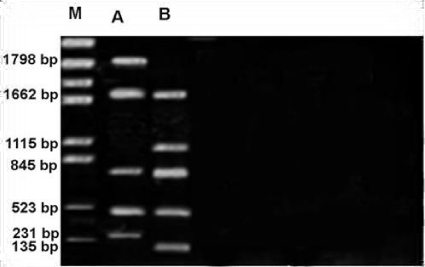
RAPD profile obtained by OPF-09 primer is depicted in the Figure 11. The size of the amplified products ranged from 164-1892bp. This profile has some differences with others such as number of monomorphic bands that it was three with 164,694 and977bp, the polymorphic was also three bands. The smallest fragment belongs to both members and Metapenaeus affinis species had the biggest one. RAPD profile obtained by OPG-16 primer is depicted in the Figure 12. The size of the amplified products ranged from 135-1798bp. Unknown member had smallest whereas the biggest one observed in Metapenaeus affinis. Of the seven fragments scored, three were monomorphic (523,845 and 1662bp) shared by both species and four fragments were polymorphic and shared by both species.
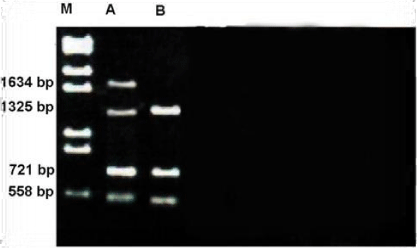
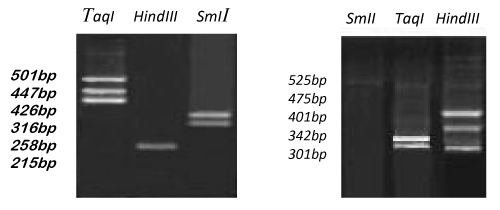
The number and size of the amplified products obtained by OPG-17 primer is represented in the Figure 13. The size of the amplified products ranged from 558-1634bp. The smallest fragment of belongs to both members Metapenaeus affinis and unknown member and the biggest one observed in Metapenaeus affinis. Four fragments were scored, of the three were monomorphic (558,721 and 1325bp) and shared by both species. Only one fragment was polymorphic in Metapenaeus affinis species. Genetic diversity was (high) between two members of Metapenaeus genus from different populations, these results indicate that although penaeids share onsiderable external features, the genetic heterogeneity at the DNA level was high in the two organisms by using RAPD-PCR. After voucher DAAPV F7 in 16s rRNA gene amplification, the segment of 16s rRNA (517bp) for both Metapenaeus affinis and suspected shrimp were digested with three suitable restriction enzymes (TaqI , SmII & HindIII ). TaqI restriction enzyme was cut the gene of Metapenaeus affinis at three sites, at 501bp, 447bp & 426bp while SmII restriction enzyme was cut it at two sites at 316bp and at 258bp whereas HindIII restriction enzyme cut the gene at only one site at 215bp. Size pieces which were cut result in differing size and differing numbers of patterns whereas another species which suspected to be Metapenaeus affinis morphologically gave different results, (TaqI )restriction enzyme cut the gene at two sites 342bp & 401bp while SmI restriction enzyme uncut any site of the gene whereas HindIII cut voucher DAAPV F7 in 16s rRNA segment at three sites at 525bp, 475bp & 301bp. Restriction patterns of TaqI, SmI & Hind III digested voucher DAAPV F7 in 16S rRNA gene clearly differentiate both Metapenaeus affinis and suspected one, as shown in the Figure 15. Overlapping patterns between different species should not be observed. Both molecular markers (RAPD and RFLP/PCR) were facilitated the task of distinguishing species that are morphologically close species and very useful in determining the distribution of the exotic species.
Conclusions
Metapenaus species were commonly found in the marine water ecosystem. In the current study, a morphologically similar but smaller freshwater version of Metapenaeus species discovered. To understand the genetic diversity and potential source of evolutionary diversion, RAPD analysis was used then it supported by RFLP-PCR marker by using 16s rRNA mitochondrial gene. Rapid Amplified Polymorphic DNA used in the genetic diversity characterization of the organism due to its low cost and ease of use. RAPD analysis gives more accurate estimates between closely related populations and less accurate estimates for distantly related populations. RAPD data used for phylogenetic studies and generally supported existing taxonomies based on morphology, isozymes and RFLPs. Conclude from RAPD & RFLP-PCR analysis evidence of genetic mismatch between the two organisms. voucher DAAPV F7 in 16S rRNA gene clearly differentiate both Metapenaeus affinis and suspected one, using RFLP-PCR molecular marker with three specific restriction enzymes as well as Genetic diversity was (high) between two members of Metapenaeus genus from different populations by using RAPD/PCR molecular marker, this present study results indicate that the two organisms are not from the same species.
This work was supported by Marine Science Centre. University of Basra, Ministry of Higher Education and Scientific Research of Iraq.
Compliance with Ethical Standards
Funding: This study was personal funded.
Ethical approval: All applicable international, national and institutional guidelines for the care and use of animals were followed.
- Ebiamadon AB, Constance ON, Nkachukwu CP (2017) Characterization and Selection of Exploitable Genetic Diversity in Soursop (Annona Muricata Linn.) Accessions Based on Phenotypic Attributes and RAPD Markers. Agroforestry Systems 4: 781-793. Link: https://bit.ly/3deMmnn
- Kapoor N (2014) Characterization of Genetic Diversity of Poplar in Three States of North India Using Rapid Primers. Int J Pharm Biol Chem Sci 4: 856-860. Link: https://bit.ly/2QuVXwT
- Thanh N (2015) Genetic Diversity of the Cultured Giant Freshwater Prawn (Macrobrachium rosenbergii) in China Based on Microsatellite Markers. Biochem Syst Ecol 59: 144-154. Link: https://bit.ly/2wn9PST
- Chen C (2017) Molecular Characterization and Genetic Diversity of Different Genotypes of Oryza Sativa and Oryza Glaberrima. Electronic J Biotechnol 30: 48-57. Link: https://bit.ly/3ahxxil
- Ibrahim A, Bakir MA, Khan HA, Al Farhan AH, Al Homaidan AA, et al. (2010) A Brief Review of Molecular Techniques to Assess Plant Diversity. Int J Mol Sci 11: 2079-2096. Link: https://bit.ly/33BUASq
- Nehemiah A, Marc K (2017) Reduced Genetic Diversity and Alteration of Gene Flow in a Fiddler Crab Due to Mangrove Degradation. Plos ONE 12: 1-20. Link: https://bit.ly/2U8oDhr
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphism's amplified by arbitrary primers and useful on genetic markers. Nucleic Acids Res 18: 6531-6535. Link: https://bit.ly/2xetbtG
- ltukhov P (2006) Intraspecific Genetic Diversity: Monitoring, Conservation, and Management. Springer. Link: https://bit.ly/2UqO5gQ
- Song H, Jung J, Kim WS (2016) Genetic Variation in a Freshwater Prawn Species, Palaemon Paucidens, in South Korea. Biochemical Systematics and Ecology 65: 23-32. Link: https://bit.ly/2UiP3Ma
- Chareontawee K, Poompuang S, Nakorn UN, Kamonrat W (2007) Genetic Diversity of Hatchery Stocks of Giant Freshwater Prawn (Macrobrachium rosenbergii) in Thailand. Mahoney, Conner. Genetic Diversity. Nova Science Publishers, Inc., 2009. Aquaculture 271: 121-129. Link: https://bit.ly/2UqMSpO
- Elibol C, Behiye B (2017) Genetic Diversity and Molecular Characterization of Natural Pancratium Maritimum L. Populations by DNA Markers. Turkish Journal of Botany 41: 569-578. Link: https://bit.ly/2wnaR1f
- Thanh N, Qigen L, Liangjie Z, Hong Z, Jun L, et al. (2015) Genetic Diversity of Cultured Populations of Giant Freshwater Prawn (Macrobrachium rosenbergii) in China Using mtDNA COI and 16S rDNA Markers. Biochem Syst Ecol 261-269. Link: https://bit.ly/3b91TU7
- Rumisha C, Leermakers M, Elskens M, Mdegela RH, Gwakisa P, et al. (2017) Genetic Diversity of the Giant Tiger Prawn Penaeus Monodon Trace Metal Pollution at the Tanzanian Coast. Marine Pollution Bulletin 114: 759-767. Link: https://bit.ly/2xS233L
- Franco J, Julian A (2012) Genetic Diversity: New Research. Nova Science Publishers, Inc. Link: https://bit.ly/392NOWU
- Thaewnon-ngiw B, Klinbunga S, Phanwichien K, Sangduen N. Lauhajinda N, et al. (2004) Genetic diversity and molecular markers of introduced and native apple snails (Genera Pomacea and Pila) in Thailand. J Biochem Mol Biol 37: 493-502. Link: https://bit.ly/2wqSPLo
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence weighting, position-specific gap penalties and weight metric choices. Nucleic Acids Res 22: 4673-4680. Link: https://bit.ly/2Wr2n3P
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. MolMar Biol Biotechnol 3: 294-299. Link: https://bit.ly/2x9R9WL
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, USA.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
