Open Journal of Bacteriology
New Methods to Remove Rhizoplane Bacterial DNA of Banana
Miaomiao Yin, Mingyue Wang, Hongming Tan and Lixiang Cao*
Cite this as
Yin M, Wang M, Tan H, Cao L (2017) New Methods to Remove Rhizoplane Bacterial DNA of Banana. Open J Bac 1(1): 016-020. DOI: 10.17352/ojb.000004The aim of this study was to evaluate the effects of different surface sterilization protocols on retained rhizoplane bacterial DNA of banana. Viable rhizoplane bacteria and bacterial DNA copies of banana roots were treated with four sterilization agents: 75% ethanol and sodium hypochlorite solution (5% available chlorine), chlorine dioxide (0.2%), peroxyacetic acid (0.4%), and formaldehyde (36%) with different incubation times. Retained viable bacteria and bacterial DNA of banana roots treated were compared based on viable count, and qPCR and MPN-PCR methods. Root treatments with peroxyacetic acid (0.4%) and formaldehyde (36%) for 5 min could remove most (99.9999%) of viable rhizoplane bacteria. Chlorine dioxide (0.2%), peroxyacetic acid (0.4%), and formaldehyde (36%) could remove 99.9% of bacterial DNA segments of 16S rRNA gene, whereas, formaldehyde (36%) could remove 99.99% of whole 16S rRNA genes of rhizoplane bacteria. The surface sterilization protocol that included incubation with formaldehyde (36%) for 9 min and further treatment with 0.1 mol l-1 NaOH for 10 min might be an effective sterilization method to remove rhizoplane bacterial rRNA genes in the study of endophytic bacterial communities of banana roots.
Introduction
Endophytes are conventionally defined as bacteria or fungi that reside internally in plant tissues, can be isolated from the plant after surface disinfection and cause no negative effects on plant growth (i.e., they are either beneficial or commensal) [1]. Endophytes are considered as a subset of the root microbiota. The composition of the root microbiota can affect important plant traits, such as stress tolerance, productivity, and fitness [2]. Although the importance of the rhizosphere microbiome for plant growth has been widely recognized, the vast majority of rhizosphere microorganisms (including endosphere microbiota) are still poorly understood [3].
Until recently, most studies on endophytes involved first isolating organisms into pure culture to identify them, the species that do not grow or grow very slowly in culture media are overlooked [4]. By the culture-dependent methods, surface sterilization was the critical step to isolate endophytes from plant tissues [5, 6]. Molecular-based methods overcome the culturability problem of many microbes by analyzing the PCR-amplified DNA products from total DNA extracted from plant tissues [7]. However, rhizosphere bacterial cells tightly attach to plant cells, so the bacterial DNA extracted from plant tissues may be contaminated by surface bacteria [8]. In order to avoid the isolation of surface bacterial DNA, potato stems and tubers were peeled asceptically [9]; however, it was not possible to peel tiny roots, mechanical removal of rhizoplane bacterial populations using vigorous shaking with glass beads had been demonstrated previously, however, the removal efficiency of rhizosphere bacterial DNA was not demonstrated [8]. Reliable surface sterilization methods are needed to be developed for analysis of endosphere microbiota.
Hypochlorite is known to be a very effective killer of bacteria, which is partly attributable to lethal DNA damage [6]. Hypochlorites have been widely used to surface sterilize plant samples to remove viable bacteria [7, 10-16]. Nevertheless, the removal efficiency of rhizosphere bacterial DNA by surface sterilization protocols was not demonstrated. After surface sterilization, it is possible that some residual DNAs are still remained on the root surfaces and might be misconsidered as endophytic bacteria by molecular-based methods.
To evaluate the efficiency of surface sterilization methods to remove rhizoplane bacterial DNA, different surface sterilization methods (hypochlorite, chlorine dioxide, peroxyacetic acid, formaldehyde) were proposed to sterilize banana roots and the residual DNAs were quantified by real-time PCR and MPN-PCR in the study.
Materials and Methods
Sample Collection
Healthy roots of banana plants (Musa sp., AAA, Cavendish subgroup, cv. Williams) were collected from a banana plantation in the suburbs of Guangzhou, China. Three banana plantlets were dug out carefully to ensure that maximal amount of root materials was collected. The samples were placed in plastic bags and taken to the laboratory and processed within 4 h of collection.
Surface Sterilization
The root samples were washed with tap water to remove soil particles and sterilized by the following protocols: (1) sequential immersion in 75% (v/v) ethanol for 5 min, and sodium hypochlorite solution (5% available chlorine) for 5 min; (2) immersion in 0.2% chlorine dioxide solution for 5 and 7 min, respectively; (3) immersion in 0.4% peroxyacetic acid solution for 5 and 7 min, respectively; (4) immersion in 36% formaldehyde solution for 5 and 7 min, respectively. Then, the sterilized roots were rinsed three times with demineralized sterile water (vortex for 2 min per rinse) to remove the surface sterilization agents. To validate the surface sterilization, the banana roots soaked with Pseudomonas putida CGMCC 1.2309 China General Microbiological Culture Collection Center (2.0×109 cfu ml-l) were treated by the above surface sterilization protocols. The sterilized banana roots were further treated with NaOH (0.1 mol l-1, 10 min), NaHCO3 (10%, 10 min), or DNase (20 mg l-1, 10 min) to remove retained bacterial DNA derived from dead cells.
Measurements of Rhizosphere Bacteria and Bacterial DNA
Surface sterilization of the banana roots was checked: (1) by rolling the sterilized roots onto PDA (Potato Dextrose Agar) plates, which were incubated for 7 days at 28 °C; (2) aliquots (0.1 ml) of the demineralized sterile water used in the final rinse were plated onto PDA plates and the plates were incubated at 28 °C for 7 days; (3) aliquots (10 ml) of the sterile water used in the final rinse were centrifugated (8000 rpm, 5 min at 4 °C) to collect bacterial cells. The bacterial DNA was extracted using E.Z.N.A. Bacterial DNA Kit (Omega) according to the manufacture’s instruction. The bacterial DNA copy numbers were further quantified by real-time PCR and MPN (most probable number)-PCR.
Amplification and detection of DNA by real-time PCR were performed with LightCycler 480 (RoChe) using optical grade 96-well plates [17]. Triplicate samples were routinely used for the determination of DNA by real-time PCR. The PCR reaction was performed in a total volume of 10 μl using the THUNDERBIRD SYBR qPCR Mix (TOYOBO), containing 0.2 μl of each of the universal forward and reverse primers (Eub338: ACTCCTACGGGAGGCAGCAG and Eub 518: ATTACCGCGGCTGCTGG) and the fluorogenic probe. The reaction conditions for amplification of DNA were 95°C 30 s, and 40 cycles of 95°C for 5 s and 60°C for 20 s. Data analysis was conducted using LightCycler480 Software 1.5 supplied by RoChe.
The bacterial DNA templates were serially diluted and the bacterial primer pairs (27F: GAGTTTGATCACTGGCTCAG and 1492R: TACGGCTACCTTGTTACGACTT) were used for multiplex PCR amplification [18]. According to the results of agarose gel electrophoresis, the samples were analyzed by the absence-presence method, with parallel analysis of five repetition tubes. Tubes were considered positive for PCR assays lead to positive results. MPN values were derived from standard tables and converted into base 10 logarithms for statistical analysis.
Extraction of total DNA from surface sterilized banana roots and amplification
The total DNA of banana roots sterilized with formaldehyde (36%) for 9 min and 0.1 mol l-1 NaOH solution for 10 min were extracted using E.Z.N.A. Plant DNA Kit (Omega) according to the manufacture’s instruction. The bacterial primer pairs (27F: GAGTTTGATCACTGGCTCAG and 1492R: TACGGCTACCTTGTTACGACTT) were used to amplify bacterial whole 16S rRNA genes according to the above methods.
Results
Sterilization with 75% ethyl alcohol did not reduce the number of viable bacteria on the rhizoplanes, further immersion in sodium hypochlorite solution (5% available chlorine) for 5 min removed 99% of rhizoplane bacteria. Chlorine dioxide (0.2%) removed 99.9% of rhizoplane bacteria after immersed for 5 min, extending treatment time to 7 min did not remove more bacteria [Table 1]. However, the chlorine dioxide only retarded the bacterial growth.
The number of bacterial colony was 33 ~ 83 cfu ml-1 when the plates were incubated for 48 hours, and more bacterial colonies (137 ~ 330 cfu ml-1) appeared when the plates were incubated more than 72 hours. Peroxyacetic acid (0.4%) and formaldehyde (36%) removed most (99.9999%) of rhizoplane bacteria when the roots were treated for 5 min. The 7 min treatment did not enhance the sterilization efficiency. The similar sterilization efficiency was further validated by rolling the roots on PDA plates [Table 2].
However, more viable bacteria on roots were found from the procedure. Therefore, the procedure was a sensitive method for validating surface sterilization. The two methods validated the effectiveness of sterilization with peroxyacetic acid (0.4%) and formaldehyde (36%), thus the rhizoplane bacterial DNA was further quantified.
The number of bacterial DNA copies in sterile water used in the final rinse determined by real-time PCR is one order of magnitude higher than that determined by plate counting [Table 3].
Sterilization with 75% ethyl alcohol removed 90% of bacterial DNA copies on the rhizoplanes, further immersion in sodium hypochlorite solution (5% available chlorine) for 5 min removed 99% of bacterial DNA. Chlorine dioxide (0.2%) did not remove viable bacteria like peroxyacetic acid (0.4%) and formaldehyde (36%). However, chlorine dioxide (0.2%), peroxyacetic acid (0.4%), and formaldehyde (36%) removed 99.9% of bacterial DNA copies. Bacterial DNA copies (1-7 ×103) were remained on the rhizoplane. Formaldehyde (36%) showed the best removal efficacy and 99.99% of bacterial whole 16S rRNA genes were removed [Table 4].
Although formaldehyde (36%) could remove 99.99% of bacterial whole 16S rRNA genes, the remained bacterial DNA could still be amplified by PCR. NaOH, NaHCO3 and DNase was further used to remove rhizoplane bacterial DNA [Table 5].
Compared with negative controls, the Ct value of retained bacterial DNA after sterilization with formaldehyde (36%) for 9 min and further treatment with 0.1 mol l-1 NaOH solution for 10 min is similar to negative control. The protocols could remove the disturbance from surface bacterial DNA of banana roots and could be used to study the plant endosphere microbiome.
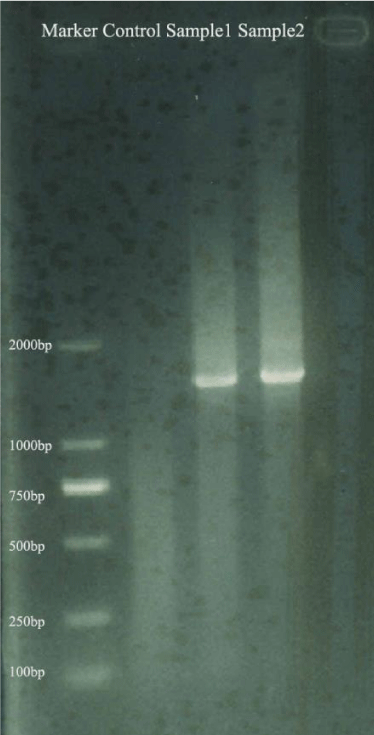
The total DNA was extracted from banana roots sterilized with formaldehyde (36%) for 9 min and 0.1 mol l-1 NaOH solution for 10 min and amplified for bacterial whole 16S rRNA genes. The whole 16S rRNA genes (1.5 kb) could be amplified from two banana root samples [Figure 1]. So the DNA extracted from surface sterilized banana roots could be used to analyze bacterial community by high-throughput sequencing technologies based on PCR (such as pyrosequencing and Illmina-based analysis).
Discussion
The microbiota colonizing the rhizosphere and the endophytic compartment contribute to plant growth, productivity, carbon sequestration, and phytoremediation. However, genetic principles governing the derivation of host-specific endophyte communities from soil communities are poorly understood [19]. The use of DNA-based metagenomic sequencing for analyzing endophytic bacterial communities has revealed that the excess bacterial cells mostly represent uncultured bacteria [20]. So the errors derived from rhizoplane bacteria should be excluded. However, the effects of dead rhizoplane bacterial DNA on the endophytic communities have not been demonstrated. Although the rhizoplane bacteria were killed by sterlization agents, the bacterial DNA could be amplified by PCR with primers towards part of or whole 16S rRNA genes. Some rhizoplane bacteria might be misconsidered as endophytic bacteria in previous reports [7, 10-16]. The endophytic bacterial communities in previous reports might contain some rhizoplane bacteria.
In the study, although peroxyacetic acid (0.4%) and formaldehyde (36%) removed 99.9999% of viable bacteria on root surface, the numbers of remained DNA copies on roots quantified by real-time PCR and MPN-PCR were about 103 and 102, respectively. The difference might derive from different quantification methods. It was speculated that formaldehyde (36%) could degrade whole 16S rRNA gene (1500 bp) into different segments, which could not be quantified by MPN- PCR towards whole 16S rRNA gene (1500 bp) but could be quantified by real-time PCR towards parts of 16S rRNA gene (180 bp). Peroxyacetic acid (0.4%) could not degrade whole 16S rRNA gene, the numbers of DNA copies by real-time PCR and MPN-PCR were both about 103. Although formaldehyde (36%) could remove 99.99% of bacterial whole 16S rRNA genes, the remained bacterial DNA could be amplified by PCR. The NaOH and DNase were used to remove bacterial DNA derived from dead cells. NaOH and DNase could remove residual bacterial DNA efficiently. After treatment with 0.1 mol l-1 NaOH solutions and DNase, the DNA copy number declined to that in negative control solution (no DNA added). The whole 16S rRNA gene could still be amplified from total DNA extracted from surface sterilized banana roots. The results showed that surface sterilization protocols could not affect the analysis on endophytic bacterial communities from banana roots by sequencing.
Compared with peroxyacetic acid (0.4%), formaldehyde (36%) is stable and low cost, so formaldehyde (36%) was suggested to sterilze the banana roots. DNase was exluded for retained DNase might degrade total DNA extracted from surface sterilized banana roots. NaOH was used to remove rhizoplane bacterial DNA due to its low price and stability.
Conclusions
In general, the killed rhizoplane bacterial DNA could still be amplified by PCR with primers towards part of or whole 16S rRNA genes. Some rhizoplane bacteria might be misconsidered as endophytic bacteria by PCR-based sequence analysis. Formaldehyde (36%) could degrade whole 16S rRNA gene (1500 bp) into different segments, however, the remained bacterial 16S rRNA gene segments could be amplified by PCR. Addition of NaOH or DNase could further remove residual rhizoplane bacterial DNA efficiently. The surface sterilization protocols could not affect the analysis on endophytic bacterial communities from banana roots by PCR-based sequencing. The protocol that sterilizion by formaldehyde (36%), and further treatment with 0.1 mol l-1 NaOH solution could be used to remove rhizoplane bacterial DNA in the studies on plant microbiomes.
- Gaiero JR, McCall CA, Thompson KA, Day NJ, Best AS, et al (2013) Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am J Bot 100: 1738-1750. Link: https://goo.gl/gs39WG
- Kristin A, Miranda H (2013) The root microbiota-a fingerprint in the soil? Plant Soil 370: 671-686. Link: https://goo.gl/tJXkcj
- Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634-663. Link: https://goo.gl/DFDXKR
- Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49: 291-315.
- Link: https://goo.gl/V6ENGk
- Schulz B, Wanke U, Draeger S, Aust HJ (1993) Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol Res 97: 1447-1450. Link: https://goo.gl/IdAwDg
- Miché L, Balandreau J (2001) Effects of rice seed surface sterilization with hypochlorite on inoculated Burkholderia vietnamiensis. Appl Environ Microbiol 67: 3046-3052. Link: https://goo.gl/fhmt6H
- Manter DK, Delgado JA, Holm DG, Stong RA (2010) Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb Ecol 60: 157-166. Link: https://goo.gl/iEQpCA
- Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, et al (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant-Microbe Int 25: 28-36. Link: https://goo.gl/Z8WvZm
- Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 39: 23-32. . Link: https://goo.gl/krdOFG
- Araújo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JWL, et al (2002) Diverisity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68: 4906-4914. Link: https://goo.gl/1LuxaJ
- Dong Y, Iniguez AL, Ahmer BMM, Triplett EW (2003) Kinetics and strain specificity of rhizosphere and endophytic colonizatio by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl Environ Microbiol 69: 1783-1790. Link: https://goo.gl/Lj9m4r
- Conn VM, Franco CMM (2004) Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl Environ Microbiol 70: 1787-1794. . Link: https://goo.gl/kvxO4d
- Chi F, Shen SH, Cheng HP, Jing XX, Yanni YG, et al (2005) Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71: 7271-7278. Link: https://goo.gl/KhvclT
- Götz M, Nirenberg H, Krause S, Woletrs H, Draeger S, et al (2006) Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol Ecol 58: 404-413. Link: https://goo.gl/3y1iCG
- Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99: 185-206. Link: https://goo.gl/DXMoks
- Li JH, Wang ET, Chen WF, Chen WX (2008) Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang Province of China. Soil Biol Biochem 40: 238-246. Link: https://goo.gl/mj9jve
- Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148: 257-266. Link: https://goo.gl/NaKw4D
- Luan X, Chen J, Liu Y, Li Y, Jia J, et al(2008) Rapid quantitative detection of Vibrio parahaemolyticus in seafood by MPN-PCR. Curr Microbiol 57: 218-221. Link: https://goo.gl/ymfnR4
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86-90.Link: https://goo.gl/ZqSW7L
- Hirsch PR, Mauchline TH (2012) Who’s who in the plant root microbiome? Nat Biotechnol 30: 961-962. Link: https://goo.gl/u0OdAV

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
