To Find out the Essentiality of Rv0526 Gene in Virulence of Mycobacterium tuberculosis by using In silico Approaches
CSIR-Institute of Genomics and Integrative Biology, Council of Scientific and Industrial Research,Mall Road, Delhi, India
Author and article information
Cite this as
Shivangi, Beg A, Meena S, Meena LS (2017) To Find out the Essentiality of Rv0526 Gene in Virulence of Mycobacterium tuberculosis by using In silico Approaches. Open J Bac. 2017; 1(1): 013-015. Available from: 10.17352/ojb.000003
Copyright License
© 2017 Yin M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Tuberculosis has emerged as a major world health problem, with almost one-third of the world population today infected with Mycobacterium tuberculosis H37Rv (M. tuberculosis). This gram-positive bacterium makes so many complications in its eradication completely. We need a proper way to inhibit its pathogenesis. Rv0526 (651bp/ 216 aa) is predicted to be a thioredoxin (Trx) like protein of Rv3673 family form a single transmembrane helix and show similarity to other M. tuberculosis thioredoxin-like proteins Rv1470, Rv1471, Rv1677. Trx are proteins that act as antioxidants by facilitating the reduction of other proteins by cysteine thiol-disulfide exchange, and it acts by its specifi c motif Cys37-Cys40 (CXXC). By using several bioinformatics tools, we may predict that Rv0526 may be an important component of cytochrome c maturation complex and show interaction with different other proteins. After knowing such specifi c features of this gene we may predict that mutational studies in Rv0526 gene may open new way towards the drug development.
M. tuberculosis: Mycobacterium tuberculosis; HIV: Human immunodeficiency virus; MDR: Multi drug resistant; XDR: Extensively drug resistant; TDR: Total drug resistant ; bp: Base pair; aa : amino acid; kD: kilo Dalton; Trx: Thioredoxin; ROS: Reactive oxygen species ; CXXC: Cystein XX Cystein ; CCM: Cytochrome c maturation
Introduction
Tuberculosis (TB) has emerged as a major world health problem, with almost one-third of the world population today infected with Mycobacterium tuberculosis H37Rv (M. tuberculosis) [1]. It is an infectious disease caused by the gram-positive bacilli M. tuberculosis [2]. This bacterium typically affects the lungs (pulmonary TB) but can also affect other sites (extra-pulmonary TB). The disease is spread by airborne Mycobacterium particles. Overall, a relatively small proportion (5–15%) of the estimated 2–3 billion people are infected with M. tuberculosis will develop TB disease during their lifetime. However, the probability of developing TB disease is much higher among people infected with HIV (Human immunodeficiency virus) [3]. Moreover, because of coinfection with HIV, non-adherence to the prescribed regimen, improper and incomplete treatment, and lack of proper diagnostic techniques have led to the emergence of multiple-drug-resistant strains. Years of investigation and genomic research find a collection of novel drugs which may help in the treatment of different varieties of TB such as MDR (multi drug resistant), XDR (extensively drug resistant) and TDR (total drug resistant). These new varieties of tuberculosis trigger us to move to form modern drugs [4-5]. This bacterium is an opportunistic pathogen of alveolar macrophages. Immunologically compromised person can get infected by this bacterium very sharply [6]. M. tuberculosis is an obligate aerobe, non-motile, slow growing (~24-hour doubling time) and nonspore forming bacteria [7]. Due to its complex nature of cell wall macrophage cannot completely eradicate the bacterium and also this bacilli inhibit fusion between phagosome and lysosome [8]. In this article, we are dealing with specific features of Rv0526 by using in silico approaches. It is predicted that it may act as an important gene of thioredoxin protein family.
In silico approaches of Rv0526
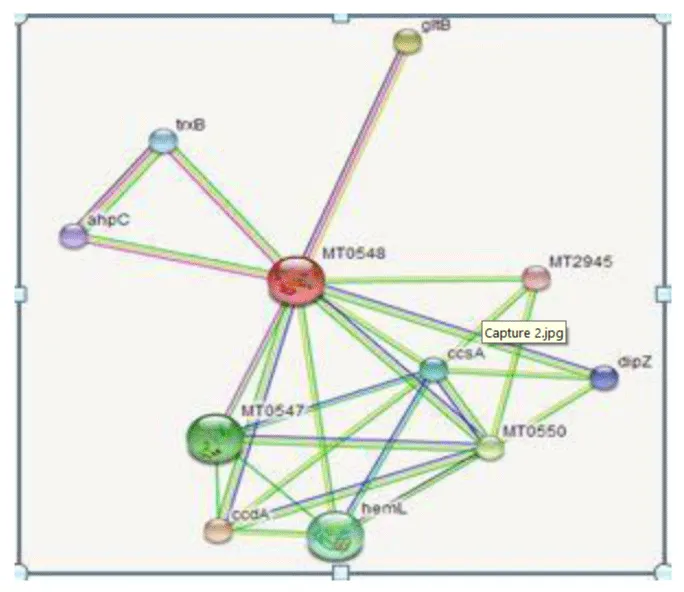
In this study, we focused towards a gene named Rv0526 (651bp/ 216 aa) having molecular weight of 23.218 kD (from nucleotide sequence). It is possibly work as Trx like protein (thiol-disulfide interchange protein) which is similar to other well defined Trx like proteins of M. tuberculosis Rv1470, Rv1471, Rv1677 [9]. Trx proteins act as antioxidants by facilitating the reduction of other proteins by cysteine thiol-disulfide exchange. Trx is found in nearly all known organisms and is essential for life in mammals [10-11]. Trx plays a critical role in the regulation of cellular redox homeostasis. Trx is able to reduce a variety of target substrates and reactive oxygen species (ROS) through the cyclization of its active site dithiol to the oxidized disulphide Cys37-Cys40 (CXXC) [12]. Rv0526 is predicted to be secreted lipoproteins. It is located in the genomic region and predicted to be contains motif, which is required for cytochrome c maturation (CCM). By using bioinformatic approach, we get different findings about this gene, which may help in getting more information about this gene to predict its functionality. Tuberculist amino acid sequence shows that Rv0526 gene containing the thioredoxin motif CXXC [13]. String database server shows the interaction of this protein with trxB, gltB, ahpC, ccsA, ccdA, hemL, dipZ, MT0550, MT2945 and MT 0547. It reveals that Rv0526 extremely interact with trxB, gltB and ahpC which prefigure that it may be involved a thioredoxin reductase [14]. As shown in Figure 1. The HMMTOP transmembrane topology prediction server predicts both the localization of helical transmembrane segments and the topology of transmembrane protein. By using HMMTOP server it is identified that gene Rv0526 forming a two transmembrane helices forming between 17-33 and 158-176 amino acid sequence, which localized in the membrane [15].
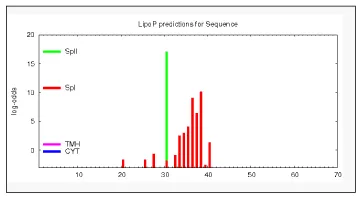
PSORT (Prediction of Protein Sorting Signal and Localization Sites in Amino Acid Sequence) analysis shows that Rv 0526 gene majorly resides in mitochondria which further predict that it is essentially involved in cytochrome C maturation and electron transport chain as shown in Table 1 [16]. Lipo P tools which help in discriminating between lipoprotein signal peptides. Lipo P confirms that Rv0526 contains SpI: signal peptide (signal peptidase I) and SpII: lipoprotein signal peptide (signal peptidase II) as shown in Figure 2 [17]. Overall result prediction for Rv0526 in our study is summaries in Table 2.
Summary
We can summarize that Rv0526 might be a Trx like protein, which is important for CCM. CCM is the significant component of an electron transport chain of aerobic respiration. Rv0526 containing CXXC motif that is responsible for the activity of Trx like proteins. It is also shows homology with Rv3673 which is proven to be act like a thioredoxin. Understanding of these specific features of Rv0526 gene conclude that this gene may act as an essential drug target. Mutation in the Cystein of this motif which is a part of Trx motif can play a significant role in killing the pathogenesis of mycobacterium. Further study required to understand the physiological and biochemical role of this gene to may develop antituberculosis drugs.
The authors acknowledge the financial support from OLP1121 and GAP0092 of the Department of Science and Technology and Council of Scientific & Industrial Research.
- Gaiero JR, McCall CA, Thompson KA, Day NJ, Best AS, et al (2013) Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am J Bot 100: 1738-1750 . Link: https://goo.gl/gs39WG
- Kristin A, Miranda H (2013) The root microbiota-a fingerprint in the soil? Plant Soil 370: 671-686. Link: https://goo.gl/tJXkcj
- Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634-663. Link: https://goo.gl/DFDXKR
- Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49: 291-315.
- Link: https://goo.gl/V6ENGk
- Schulz B, Wanke U, Draeger S, Aust HJ (1993) Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol Res 97: 1447-1450. Link: https://goo.gl/IdAwDg
- Miché L, Balandreau J (2001) Effects of rice seed surface sterilization with hypochlorite on inoculated Burkholderia vietnamiensis. Appl Environ Microbiol 67: 3046-3052. Link: https://goo.gl/fhmt6H
- Manter DK, Delgado JA, Holm DG, Stong RA (2010) Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb Ecol 60: 157-166. Link: https://goo.gl/iEQpCA
- Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, et al (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant-Microbe Int 25: 28-36. Link: https://goo.gl/Z8WvZm
- Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 39: 23-32. . Link: https://goo.gl/krdOFG
- Araújo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JWL, et al (2002) Diverisity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68: 4906-4914. Link: https://goo.gl/1LuxaJ
- Dong Y, Iniguez AL, Ahmer BMM, Triplett EW (2003) Kinetics and strain specificity of rhizosphere and endophytic colonizatio by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl Environ Microbiol 69: 1783-1790. Link: https://goo.gl/Lj9m4r
- Conn VM, Franco CMM (2004) Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl Environ Microbiol 70: 1787-1794. . Link: https://goo.gl/kvxO4d
- Chi F, Shen SH, Cheng HP, Jing XX, Yanni YG, et al (2005) Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71: 7271-7278. Link: https://goo.gl/KhvclT
- Götz M, Nirenberg H, Krause S, Woletrs H, Draeger S, et al (2006) Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol Ecol 58: 404-413. Link: https://goo.gl/3y1iCG
- Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99: 185-206. Link: https://goo.gl/DXMoks
- Li JH, Wang ET, Chen WF, Chen WX (2008) Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang Province of China. Soil Biol Biochem 40: 238-246. Link: https://goo.gl/mj9jve
- Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148: 257-266. Link: https://goo.gl/NaKw4D
- Luan X, Chen J, Liu Y, Li Y, Jia J, et al(2008) Rapid quantitative detection of Vibrio parahaemolyticus in seafood by MPN-PCR. Curr Microbiol 57: 218-221. Link: https://goo.gl/ymfnR4
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86-90.Link: https://goo.gl/ZqSW7L
- Hirsch PR, Mauchline TH (2012) Who’s who in the plant root microbiome? Nat Biotechnol 30: 961-962. Link: https://goo.gl/u0OdAV
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
