Open Journal of Bacteriology
Illumina Based Analysis of Bacterial and Fungal Flora in Foreguts and Hindguts of Crucian Carp (Carassius aumtus) in Retail Markets
Mingyue Wang, Miaomiao Yin, Hongming Tan, and Lixiang Cao*
Cite this as
Wang M, Yin M, Tan H, Cao L (2017) Illumina Based Analysis of Bacterial and Fungal Flora in Foreguts and Hindguts of Crucian Carp (Carassius aumtus) in Retail Markets. Open J Bac 1(1): 001-006. DOI: 10.17352/ojb.000001The intestinal microbiota of fish are the major cause for spoilage. To elucidate the bacterial and fungal community of guts, the bacterial and fungal taxa in foreguts and hindguts of crucian carp (Carassius aumtus) were analyzed by Illumina-based sequencing. Results suggested that the foreguts contained more diverse bacteria than those in hindguts; nevertheless, the hindguts contained more diverse fungi than foreguts. The anaerobic bacterial genera Cetobacteri Desulfovibrio and Shewanella in foreguts were still detected in hindguts. The fungal taxa in foreguts were different from those in hindguts. The dominant fungal genera Alternaia (78.6%), Massarina (0.8%) and Fusarium (0.2%) were only detected in hindguts. It was considered that the Alternaria, Emericella, and Cochliobolus might derive from the diets of crucian carp. The activities of Desulfovibrio might be responsible for the changes in odour, flavor and texture of the fish meat. The H2S produced by Desulfovibrio is potentially a major toxin to the fish gut epithelium and promoted the fish spoilage processes. The results are helpful for manipulation of intestinal flora to preserve fresh crucian carp in tanks.
Introduction
As with good survival rate, high reproduction rate, and disease resistance, crucian carp (Carassius auratus) is widely bred across Eurasia and America [1]. In China, crucian carp is one of the most economically important freshwater-cultured fish species. The production yield reached nearly 2,000,000 tons in 2009 [2]. However, fish are more perishable than other muscle foods, and a considerable number of fish are spoiled due to lack of good preservation. In world, 30% of landed fish are spoiled and lost [3].
Fresh fish spoilage can be very rapid after it is caught [3]. Crucian carp is an easily perishable product because of its relatively high quantities of volatile basic nitrogen as well as free amino acids, high water activity, and presence of autolytic enzymes [2]. As one of the most important food fish in China, only alive crucian carp fish is accepted in market [4]. The spoilage of fish is a complicated process in which microbial, physical and chemical variations interact. Activity of the fish’s own enzymes and chemical reactions are usually responsible for the initial loss of fish freshness, whereas the metabolic activities of microorganisms are involved in the whole spoilage [2]. Gutting of the fish immediately after capture can avoid the invasion of digestive tract proteases through the abdominal cavity to the tissue and prevent or slow degradation [3]. However, there may be chances of bacterial cross-contamination of fish during the gutting procedure. Microbial growth and metabolism are the major cause for food spoilage, the microbial populations may shift during storage and only a small fraction of fish microbiota is responsible for spoilage, known as “specific spoilage organisms” [5]. These specific spoilage organisms are present in low number in fresh fish and can eventually become dominant in spoilage microorganisms [6].
Bacterial flora isolated from eggs, skins, gills, and intestines have been described for some fish species. Bacteria recovered from the skin and gills may be transient rather than resident on the fish surfaces [7]. The gastrointestinal microbiota in fish is constituted of facultative and obligate anaerobes, which may vary among fish species with different digestive apparatus [8]. However, the zebrafish intestinal habitat select for specific bacterial taxa despite radical differences in host provenance and domestication status [9]. So far, the microbial community in fish guts has not been systematically characterized [10]. Most previous studies used traditional culture-dependent or DGGE, clone library methods to investigate the fish intestinal bacterial diversity [11-17]. With the development of the next-generation sequencing technologies, high-throughput sequencing platform is available for characterizing the bacterial community in the gut of fish [10,18-20]. More comprehensive information on fish gut microbiota would be obtained by high-throughput sequencing approaches. Illumina has fewer errors than 454 sequencing and it could provide a higher phylogenetic resolution than 454 based approaches [21]. The advantage of Illumina to provide 30 times more reads would enable us to perform in depth sequencing of samples in one run, making it an excellent tool for gut microbial diversity.
The objective of this study was to elucidate the bacterial and fungal flora in foreguts and hindguts of crucian carp. Specially, total DNAs of foreguts and hindguts were purified and analyzed by Illumina-based sequencing. The bacterial and fungal communities in foreguts and hindguts were further compared, the spoilage bacterial and fungal taxa in foreguts and hindguts were elucidated in the study.
Material and Methods
Fish sample
Ten live commercial-sized crucian carp with average weight of 250 ± 20 g were purchased from aquatic market in March, 2015. They were kept alive before being processed. The fish were killed by slurry ice and gutted under sterile conditions. The portion of intestinal tract posterior from bile duct to the first distal loop (foreguts) and the intestinal tract anterior from anus to last anterior loop (hindguts) were removed for DNA extraction.
DNA Extraction
The foreguts (designated as QianC), hindguts (designated as HouC) were used for total DNA extraction. The total DNA was extracted using PowerSoil® DNA Isolation Kit (Mol Bio) according to the manufacture’s instruction. Total DNA concentration and purity were monitored on 1% agarose gels.
Amplicon Generation and Illumina MiSeq sequencing
The primers S-D-Bact-0341-b-S-17 (5’- CCTACGGGNGGCWGCAG - 3’) and S-D-Bact-0785-a-A-21 (5’ – GACTACHVGGGTATCTAATCC - 3’) targeting the V3-V4 hyper variable regions of bacterial 16S rRNA genes were selected for analyze bacterial taxa [22]. The primers ITS5-1737F: GGAAGTAAAAGTCGTAACAAGG; ITS2-2043R: GCTGCGTTCTTCATCGATGC targeting the ITS2 regions of fungal rRNA genes were adopted to analyze fungal taxa [23]. Both forward and reverse primers were tagged with adapter, pad and linker sequencing. Each barcode sequence was added to the reverse primer for pooling multiple samples into one run of sequencing. All PCR reactions were performed in a total volume of 30μL containing 15μL Phusion® High-Fidelity PCR Master Mix (New England Biolabs) and 0.5 units of AccuPrimer TM Taq DNA Polymerase (Life Technologies, USA), 0.2 μM of forward and reverse primers, and 10 ng template DNA. Thermal cycling conditions were as follows: an initial denaturation at 98 ºC for 1 min, each of 30 cycles at 98 ºC for 10 s, 50 ºC for 30 s, and 72 ºC for 60 s, with a final extension at 72 ºC for 5 min.
Following amplification, 2 μL of PCR product was used to verify successful amplification by 2% agarose gel electrophoresis. The products of triplicate PCR reaction from one sample were combined and the pooled mixtures were purified with GeneJET Gel Extraction Kit (Thermo Scientific) and analyzed on an Agilent 2100 Bioanalyzer using High Sensitivity DNA Chips (Agilent Technologies, Germany) for size distribution. The sequencing libraries were generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Germany). Finally, the library was sequenced on an Illumina MiSeq platform at Magigen biotechnology Co. Ltd, Guangzhou, China.
Combination and data preprocessing
Forward and reverse sequences were merged by overlapping paired-end reads using FLASH (V1.2.7, https://ccb.jhu.edu/software/FLASH/) [24]. All sequence reads with the same tag were assigned to the same sample according to the unique barcodes (raw tags). The raw tags were further strictly filtered by previous methods (clean tags) [25] and the quality of clean tags were detected by Qiime (V1.7.0 https://qiime.org/index.html) [22]. The low quality tags were removed. The tags with chimera were detected and removed using UCHIME Algorithm (https://www.drive5.com/usearch/manual/uchime_algo.html) [26,27]. The effective sequences were then clustered into operational taxonomic units (OTU) at 97% sequence similarity using the UPARSE-OTU and UPARSE-OTUref algorithms of UPARSE software package (Uparse v7.0.1001 https://drive5.com/uparse/), the indices of alpha diversity were calculated [28]. Finally, the RDP classifier was used to assign representative sequence to the microbial taxa [29]. Sequence data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession number SRP062743.
Statistical analysis
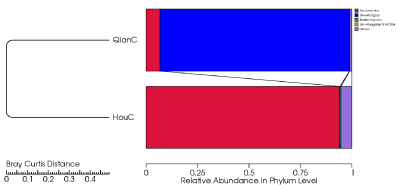
Cluster analysis was preceded by principal component analysis (PCA) using the QIIME software package. QIIME calculates both weighted and unweighted unifrac distances, which are phylogenetic measures of beta diversity [30], the phylogenetic relations among different microbial taxa were further displayed by KRONA [31]. Alpha diversity indices Chao1, ACE, Shannon, Simpson and coverage were calculated to reflect the diversity and richness of the endophytic community in different samples [32].
Results
Fungal and bacterial species richness and diversity
After qualify filtering the raw reads, 100013 bacterial sequences remained with an average length of 440 bp, 69227 fungal sequences remained with an average length of 284 bp (Table 1). The number of different bacterial OTUs at the 97% similarity level ranged from 207 to 229 per sample with an average of 221 OTU. The number of different fungal OTUs at the 97% similarity level ranged from 56 to 128 per sample with an average of 94 OTU. The alpha diversity indices (Shannon and Simpson) calculated from bacterial OTUs of different samples indicated that the foreguts contained more diverse bacteria than those in hindguts; nevertheless, the hindguts contained more diverse fungi than foreguts (Table 2).
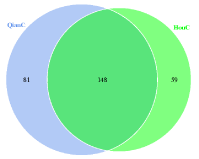
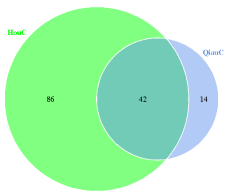
Total 229 bacterial OTUs were detected in foreguts, 148 OTUs were still detected in hindguts (Figure 1). Contrary to bacteria, the hindguts contained more fungal OTU than foreguts, and 42 OTUs were detected in foreguts simultaneously (Figure 2).
Fungal and bacterial composition and community structure
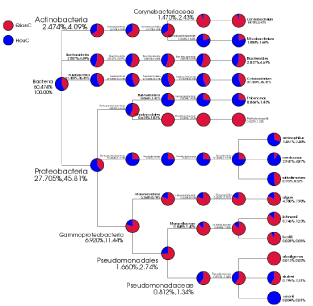
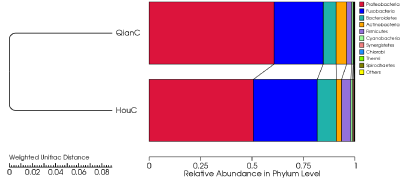
Bacterial representative sequences of each OTU were classified into the bacterial domain (99.96 % of the total data set), the Cetobacterium were the most dominant bacterial genus observed (27.5%). Desulfovibrio were the second most abundant (19.3%). Fungal representative sequences of each OTU were classified into fungi (100 % of the total data set), the Alternaria were the most dominant genus (39.3%). Emericella were the second most abundant (6.9%). The dominant bacterial and fungal species were different in the different samples. The bacterial genera Cetobacterium (23.9%), Desulfovibrio (9.3%), Shewanella (8.5%), Bacteroides (3.2%), Coryrebacterium (2.8%), Acinetobacter (1.5%) and Methyloversatilis (1.2%) were the most detected bacterial genera in foreguts, the hindguts contained Cetobacterium (31.1%), Desulfovibrio (29.3%), Shewanella (2.0%), Bacteroides (2.5%), Mycobacterium (1.6%) and Thiomonas (1.3%) as the dominant bacterial genera (Figure 3). With the digestion of food from foreguts to hindguts, the dominant bacterial genera Shewanella and Bacteroides were still detected in hindguts, other bacterial genera Pseudomonas (0.8%), Thiomonas (0.9%), Staphylococcus (0.3%), Dysgonomonas (0.4%) and Streptomyces (0.5%) were detected both in foreguts and hindguts. Dietzia (0.4%), Deinococcus (0.2%), Flavobacterium (0.1%), Chryseobacterium (0.1%), Anaerococcus (0.2%), Neisseria (0.1%), Escherichia (0.1%) and Enhydrobacter (0.1%) were detected in foreguts not in hindguts, whereas Gordonia (0.1%), Propionicimonas (0.1%) and Halothiobacillus (0.2%) were only detected in hindguts (Figure 3).
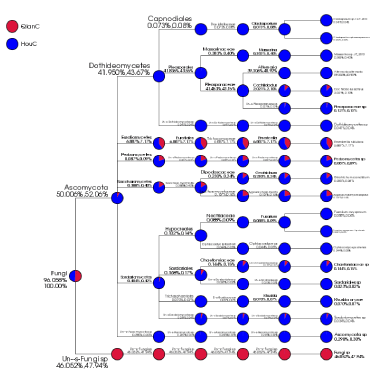
For the dominant fungal genera in the hindguts, Alternaria (39.3%), Emericella (6.9%) and Cochliobolus (2.0%) were the most detected fungal genera (Figure 4), the dominant fungal genera Emericella (6.9%) and Cochliobolus (2.0%) coexisted in foreguts and hindguts. The Zygosaccharomyces (0.1%) were detected both in foreguts and hindguts. Nevertheless, Alternaia (78.6%), Massarina (0.8%) and Fusarium (0.2%) were only detected in hindguts (Figure 4).
The bacterial phyla Proteobacteria (55.6%), Fusobacteria (27.5%) and Bacteroidetes (7.7%) in phylum level with high abundance were detected in two types of samples (Figure 5). The fungal phyla Ascomycota (50.4%) and Basidiomycota(0.3%) were detected in the two types of samples (Figure 6).
Discussion
Crucian carp is one of the most important food fish in China, and only alive fish is accepted in markets. Aquariums and tanks displaying live crucian carp caught in supermarkets and retail outlets are becoming increasingly common in China [4]. The water body in tanks is different from that in aquaculture, the effects of water body on intestinal flora of live crucian carp caught is still unknown. Our results illustrated that the foreguts contained more diverse bacteria than those in hindguts; nevertheless, the hindguts contained more diverse fungi than those in foreguts. The bacterial diversity distributed in foreguts and hindguts of crucian carp is consist with that in indigenous planktivorous gizzard shad, contratry to that in invasive Asian siler carp in Mississippi river basin, USA [10]. Although gut microbiota has become an integral component of the host, and received increasing attention [18], the fungal diversity in fish gut flora is not still reported [10]. The fish gut microbiota is only focused on bacteria [7-19]. Our results showed that the hindguts contained more diverse fungi than those in foreguts. The fungal phyla Ascomycota and Basidiomycota with high abundance were detected in guts of crucian carp. Fungal orders Pleosporales, Eurotiales, Saccharomycetales in foreguts were still detected in hindguts. The fungal taxa in foreguts were different from those in hindguts. The dominant fungal genera Alternaia (78.6%), Massarina (0.8%) and Fusarium (0.2%) were only detected in hindguts. Most Alternaria species are saprophytes that are commonly found in soil or on decaying plant tissues. Some species are opportunistic plant pathogens that cause a range of disease (stem cancer, leaf blight, or leaf spot) with economic impact on the variety of important agronomic crops [33]. Alternaria species have been frequently isolated as endophytes from leaves and stems but not from roots [34-36]. Alternaria alternate has been isolated as the most dominant species in leaves [35]. So it was considered that the Alternaria, Emericella, and Cochliobolus might derive from the diets of crucian carp.
More diverse bacteria in foreguts were detected than those in hindguts. The anaerobic bacterial genera Cetobacterium Desulfovibrio and Shewanella in foreguts were still detected in hindguts. The Cetobacterium spp. had been detected in the intestinal contents of goldfish (Carassius auratus) [8]. The Shewanella also been found in intestinal contents of goldfish, yellow catfish, rainbow rout [8,11,17]. The sulphate-reducing bacteria Desulfovibrio had not been detected in fish guts [7-19), although the Desulfovibrio are the most routinely recovered from animal and human faeces [37]. The results illustrated that the intestinal flora varied after the live crucian carp was caught and maintained in the tanks. The Desulfovibrio served as terminal oxidizers in the anaerobic degradation of organic matter entering the gastrointestinal tract and reduce sulphur and suphur-containing compounds to hydrogen sulphide (H2S) [37]. The production of H2S by the Desulfovibrio is potentially a major toxin to the fish gut epithelium and promoted the fish spoilage processes. The activities of Desulfovibrio might be responsible for the changes in odour, flavor and texture of the fish meat. Our results were consistent with the fact that fresh fish spoilage can be very rapid after it is caught [3]. So the manipulation of intestinal flora to preserve fresh crucian carp in tank would be further studied.
- Xiao J, Zou T, Chen Y, Chen L, Liu S, et al (2011) Coexistence of diploid, triploid and treraploid crucian carp (Carassius auratus) in natural waters. 12:20. Link: https://goo.gl/20KjTi
- Li T, Li J, Hu W, Zhang X, Li X, et al. (2012) Shelf-life extension of crucian carp (Carassius auratus) using natural preservatives during chilled storage. Food Chemistry 135:140-145. Link: https://goo.gl/QoBnUA
- Ghaly AE, Dave D, Budge S, Brooks MS (2010) Fish spoilage mechanisms and preservation techniques: review. American Journal of Applied Sciences 7: 859-877. Link: https://goo.gl/6ablrG
- Zeng P, Chen T, Shen J (2014) Effects of cold acclimation and storage temperature on crucian carp (Carassius auratu gibelios) in a waterless preservation. Fish Physiology and Biochemistry 40: 973-982. Link: https://goo.gl/1AZBWy
- Wang H, Luo Y, Huang H, Xu Q (2014) Microbial succession of grass carp (Ctenopharyngodon idellus) filets during storage at 4 ºC and its contribution to biogenic amines’ formation. Int J Food Microbiol 190: 66-71. Link: https://goo.gl/xOZS5Y
- Zhang Y1, Li Q1, Li D1, Liu X1, Luo Y (2015) Changes in the microbial communities of air-packaged and vacuum-packaged common carp (Cyprinus carpio) stored at 4 ºC. Food Microbiology 52: 197-204. Link: https://goo.gl/6QWiSA
- Cahill MM (1990) Bacterial flora of fishes: a review. Microbial Eology 19: 21-41. Link: https://goo.gl/9erL1x
- Silva FC1, Nicoli JR, Zambonino-Infante JL, Kaushik S, Gatesoupe FJ (2011) Influence of the diet on the microbial diversity of faecal and gastrointestinal contents in gilthead sea bream (Sparus aurata) and intestinal contents in goldfish (Carassius auratus). FEMS Microbiology Ecology 78: 285-296. Link: https://goo.gl/uX0mHo
- Roeselers G1, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, et al. (2011) Evidence for a core gut microbiota in the zebrafish. ISME J 5: 1595-1608. Link: https://goo.gl/Z8PZYp
- Ye L, Amberg J, Chapman D, Gaikowski M, Liu WT (2016) Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J 8: 2076. Link: https://goo.gl/2BdoWu
- Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, et al (2004) Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum). J Appl Microbiol 96: 117-132. Link: https://goo.gl/SvkeSr
- Al-Harbi AH, Uddin N (2005) Bacterial diversity of tilapia (Oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture 250: 566-572. Link: https://goo.gl/Qkr779
- Pond MJ, Stone DM, Alderman DJ (2006) Comparison of conventional and molecular techniques to investugate the intestinal microflora of rainbow trout (Oncorhynchus mykiss). Aquaculture 261: 194-203. Link: https://goo.gl/LRgSaj
- Ward NL1, Steven B, Penn K, Methé BA, Detrich WH 3rd. (2009) Characterization of the intestinal microbiota of two antarctic notothenioid fish species. Extremophiles 1 3: 679-685. Link: https://goo.gl/5ShtMI
- Han S, Liu Y, Zhou Z, He S, Cao Y, et al. (2010) Analysis of bacterial dicersity in the intestine of grass carp (Ctenopharyngodon idellus) based on 16S rDNA gene sequences. Aquaculture Research 42: 47-56. Link: https://goo.gl/DHFPVw
- Tapia-Paniagua ST1, Chabrillón M, Díaz-Rosales P, de la Banda IG, Lobo C, et al. (2010) Intestinal microbiota diversity of the flat fish Solea senegalensis (Kaup, 1858) following probiotic administration. Microb Ecol 60: 310-319. Link: https://goo.gl/JBF5jb
- Wu S, Gao T, Zheng Y, Wang W, Cheng Y, et al. (2010) Microbial diveristy of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 303: 1-7. Link: https://goo.gl/dh4nRw
- Wu S, Wang G, Angert ER, Wang W, Li W, et al. (2012) Composition, diversity, and origin of the bacterial community in grass carp intestine. Plos one 7: e30440. Link: https://goo.gl/gUiHCS
- Carda-Dieguez M, Mira A, Fouz B (2013) Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiology Ecology 87: 451-459. Link: https://goo.gl/TJMvXa
- Rungrassamee W1, Klanchui A1, Maibunkaew S1, Chaiyapechara S2, Jiravanichpaisal P, et al. (2014) Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon). Plos one 9: e91853. Link: https://goo.gl/AkKqbi
- Shi Y1, Yang H, Zhang T, Sun J, Lou K (2014) Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl Microbiol Biotechnol 98: 6375-6385. Link: https://goo.gl/3PhHqf
- Caporaso JG1, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621-1624. Link: https://goo.gl/yNpnrI
- Degnan PH, Ochman H (2012) Illumina-based analysis of microbial community diversity. ISME J 6: 183-194. Link: https://goo.gl/423p1X
- Magoč T1, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957-2963. Link: https://goo.gl/rT3bPJ
- Bokulich NA1, Subramanian S, Faith JJ, Gevers D, Gordon JI, et al. (2013) Quality-filtering vastly improves diversity estimates from illumine amplicon sequencing. Nature Methods 10: 57-59. Link: https://goo.gl/r5tHmA
- Edgar RC1, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194-2200. Link: https://goo.gl/G6teX1
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, et al. (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research 21: 494-504. Link: https://goo.gl/My4D1Q
- Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods 10: 996-998. Link: https://goo.gl/2PbhjK
- Wang Q1, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261-5267. Link: https://goo.gl/mHqXCA
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, et al. (2013) Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology 22: 5271-5277. Link: https://goo.gl/SMkO2D
- Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a Web browser. BMC bioinformatics 12: 385-394. Link: https://goo.gl/I0XHCm
- Kemp PF, Aller JY (2004) Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47: 161-177.1 Link: https://goo.gl/asR4N8
- Thomma BP (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol 4: 225-236. Link: https://goo.gl/e2Ac2f
- Morita S, Azuma M, Aoba T, Satou H, Narisawa K, et al. (2003) Induced systemic resistance of Chinese cabbage to bacterial leaf spot and Alternaria leaf spot by the root endophytic fungus, Heteroconium chaetospira. J Gen Plant Pathol 69: 71-75. Link: https://goo.gl/sWxqt2
- Guo LD, Xu L, Zheng WH, Hyde KD (2004) Genetic variation of Alternaria alternate, an endophytic fungus isolated from Pinus tabulaeformis as determined by random amplified microsatellites (RAMS). Fungal Diversity 16: 53-65. Link: https://goo.gl/Hs9JyX
- Gu W (2009) Bioactive metabolites from Alternaria brassicicola ML-P08, an endophytic fungus residing in Malus halliana. World J Microbiol Biotechnol 25: 1677-1683. Link: https://goo.gl/0YkP7Y
- Scanlan PD1, Shanahan F, Marchesi JR (2009) Cuture-independent analysis of Desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol 69: 213-221. Link: https://goo.gl/H4gxfB

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
