Journal of Clinical Microbiology and Biochemical Technology
Towards a fast detection of microbial resistance to antibiotics
Leonardo Venturelli1, Anne-Céline Kohler1 and Sandor Kasas1,2*
2University Center of Legal Medicine (CURML), Rue du Bugnion 9, University of Lausanne, CH-1005 Lausanne, Switzerland
Cite this as
Venturelli L, Kohler AC, Kasas S (2020) Towards a fast detection of microbial resistance to antibiotics. J Clin Microbiol Biochem Technol 6(1): 010-013. DOI: 10.17352/jcmbt.000038Introduction
The antimicrobial resistance is the ability of bacteria and fungi to proliferate even in presence of antibiotic or antimycotic drugs [1]. In the last twenty years we have been witnessing a colossal increase in the antimicrobial resistance mainly due to the abuse and misuse of these compounds [2]. Although nowadays the use of antibiotics and antimycotics is more strictly regulated, antimicrobial treatment sometimes starts before a complete identification of the infectious agent [3-5]. The diagnostic tools currently approved by the Food and Drug Administration (FDA) or European Medicine Agency (EMA) (and the other region-specific regulatory agencies) for Antimicrobial Susceptibility Test (AST) are able to provide results in a relatively slow time that can last from one day up to a month [6]. This lag time is often too long for patients suffering from rapidly evolving infections such as sepsis [7]. This is one of the main reasons why in such cases the antibiotic treatment is given to the patient before receiving AST results. Unfortunately, broad spectrum treatments may lead to resistant strains [8].
To prevent the increase of resistant strains, numerous groups developed various nanomechanical devices to perform rapid ASTs [9,10]. MEMS used in biomedical applications gained attention due to their broad applicability, scalability, and reliability [11-14]. Lab-on-a-chip and cantilever-based detection system are very promising and could lead in the next future to low cost, easy-to-use and pocket-sized AST devices. Moreover, silicon-based sensors are nowadays produced at a reasonable cost using fast standardized fabrication methods.
Nanomechanical sensors based on atomic force microscope cantilevers
Binnig and coworkers [15] developed the first Atomic Force Microscope (AFM) in 1986. The instrument consists in a tip that scans the surface of the sample. The tip is fixed at the end of a soft cantilever that deflects during the scan according to the sample’s topography. The vertical movements of the cantilever are recorded with an astonishing precision (up to 0.1 Å) and allow the reconstruction of 3D topography of the sample. Since its first iteration, the instrument improved and is now used in practically all research areas, ranging from chemistry to medicine [16,17]. Nowadays, an AFM can image biological samples in almost physiological environment at an atomic resolution, detect interaction forces between single molecules [18,19] or assess the mechanical properties of single bacteria with an unpreceded resolution. The biomedical application of cantilever-based sensors, in particular in the field of microbiology, is broad and covers different topics. It allows to precisely analyze the mechanical properties of a specific cell area and to record bacterial division at a nanometric scale [20,21]. Indeed, the microbial cells can be studied both by probing their mechanical properties under the AFM tip or by having them directly attached onto the cantilever or even inside it by the means of embedded channels [22-24].
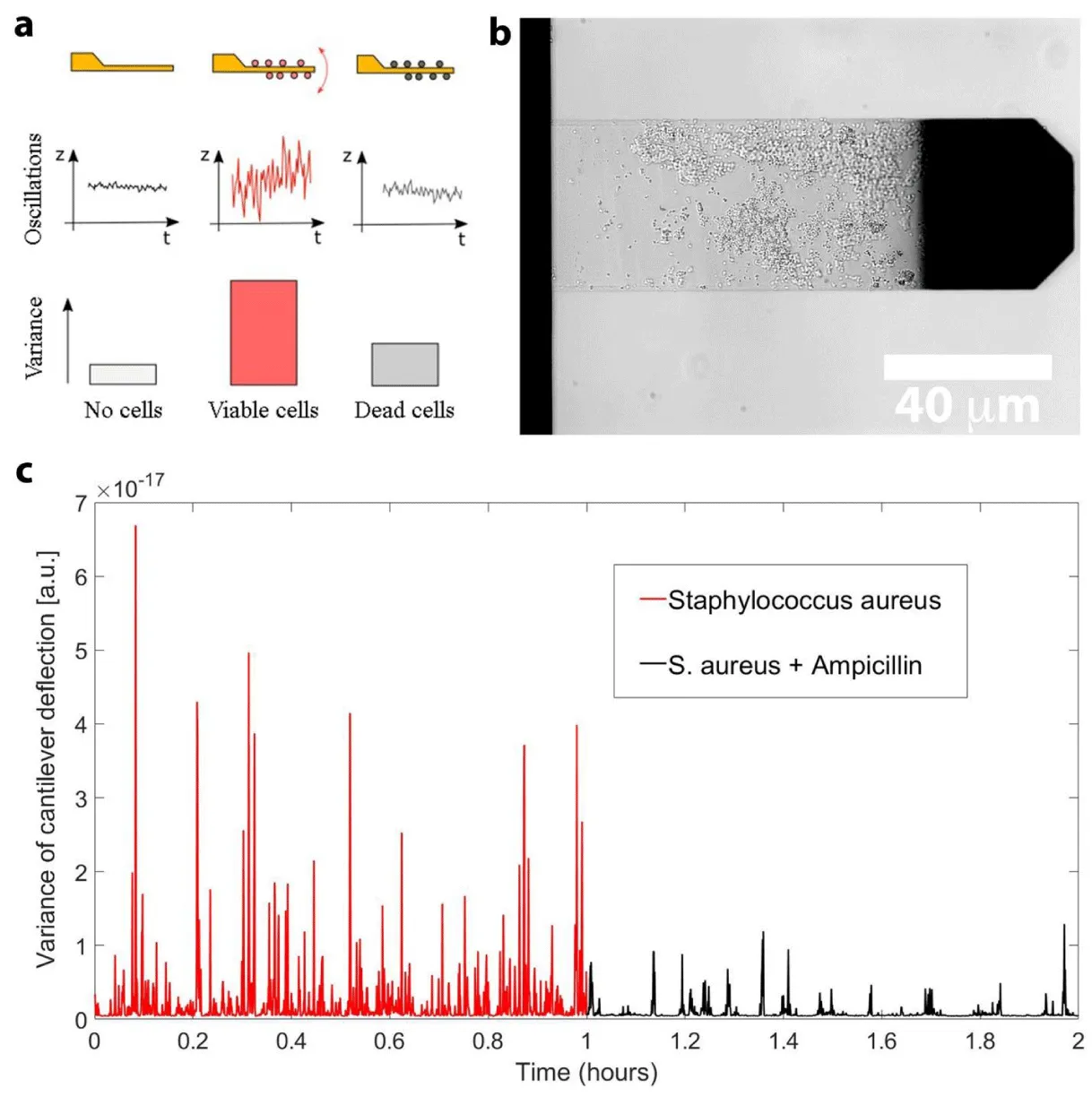
In 2013, Longo and colleagues developed a technique, referred to as nanomotion detection, where the microorganism of interest is attached onto an AFM cantilever and the cantilever oscillations are monitored as a function of time and of different chemicals [25]. The authors demonstrated that living organisms induce nanometric scale oscillations of the cantilever that immediately stop when the organism dies. Remarkably, this study revealed that the method is well suited to conduct rapid ASTs: reliable results being delivered in a timeframe of minutes to hours. Figure 1 represents the working principle of the method as well an example of a nanomotion experiment performed on Staphylococcus aureus. Briefly, the microorganism of interest is attached onto the cantilever and the oscillations of the lever are recorded for a minimum of 20 min. In a second step, the microorganism is exposed to the drug to be tested and the viability of the cells is assessed by monitoring the cantilever oscillations. Taken all the steps together, a typical experiment lasts for 1 to 3 hours, decreasing a lot the time delay of the AST.
Double blind experiments on blood culture pellets, using the nanomotion technique, demonstrated an accuracy above 95% when analyzing resistant bacteria obtained from patient suffering of infections [26]. This is one article were the accuracy of the technique was checked in a real situation, using samples obtained from patients in order to test the technique outside the academic laboratory, even though still handled by experts in the field.
Therefore, the nanomotion detection technique is extremely promising and its use as diagnostic tool is foreseeable in the close future.
By using the very same technique several studies demonstrated that all of the so far explored living organisms oscillate at a nanometric scale as long they are alive. Indeed, this novel method was successfully tested on dozens of organisms such as bacteria, yeast [27], vegetal and mammalian cells [28]. Applied on bacteria, the technique was proven to be independent on the gram staining, the motility, or the replication rate of the organism [29-31].
Similar techniques that use micro or nano-fabricated device to detect the presence of several kind of different compounds, or living organisms have been developed [32]. Their potential application covers several scientific area and it is particularly well suited for biomedical applications [33].
The main drawback of AFM-cantilever based AST is probably related to the need to attach the organism of interest onto a lever with a good reliability. The compounds used to accomplish this task are usually not widely usable in terms of applicability to several different strain or species. Indeed, as for common diagnostic AST used nowadays in clinics the culture medium changes (for instance: liquids and solids containing different nutrients) among different microorganisms, the attaching compound (needed to load the cells on the lever) should be carefully checked or tested to ensure that the microorganism, obtained from the patient, is correctly bound and identified. Therefore few biological and chemical products have been testing to load microorganisms, such as bacteria and yeasts, on cantilevers to ensure an sufficient loading capacity. For instance, some of those compounds may lead to toxic or metabolic-impairing effect on few microorganisms and thus biasing their reaction to antibiotics and antimycotics. Based on the data available from the literature, some of the aforementioned compounds are more suitable for bacteria such as glutaraldehyde, APTES, poly-L-lysine and fibronectin whereas others such as Concanavalin-A more for fungi and yeasts. Another potential issue that may limit the usability of this type of AST techniques as diagnostic tool is the relative complexity in the required steps needed to finally have an acceptable number of viable microorganisms on the cantilever. However, this last drawback can be overcome by developing automatized systems able to help the final users throughout complex steps.
The potential of these techniques pertains to the fact that due to their nano-scale sizes they offer the possibility to study microbial activity down to few or even single cell level [34-44].
Conclusion
In this work we introduce a non-exhaustive view of a novel AST technique to rapidly and accurately detect microbial resistance to antibiotics. Hopefully, in a close future, this method will lead to fast, robust and easy-to-use devices. The antibiotic resistance is indeed a worldwide treat that should be tackled as fast as possible and a credible option to achieve this goal is the use of rapid ASTs. Such devices could reduce the use of large spectrum antibiotics thus limiting the development of resistant strains.
The authors were funded by the Swiss National Grants 200021-144321 and 407240_167137, CRSII5_173863 the Gebert Rüf Stiftung GRS-024/14 and NASA NNH16ZDA001N-CLDTCH.
- Theuretzbacher U, Bush K, Harbarth S, Paul M, Rex JH, et al. (2020) Critical analysis of antibacterial agents in clinical development. Nat Rev Microbiol 1-13. Link: https://bit.ly/2VxESDU
- Alanis AJ (2005) Resistance to Antibiotics: Are We in the Post-Antibiotic Era? Arch Med Res 36: 697-705. Link: https://bit.ly/2XBUfOt
- Daniels R (2011) Surviving the first hours in sepsis: getting the basics right (an intensivist's perspective). J Antimicrob Chemother 66: ii11-ii23. Link: https://bit.ly/2K6e74f
- Farrell JJ, Hujer AM, Sampath R, Bonomo RA (2015) Salvage microbiology: opportunities and challenges in the detection of bacterial pathogens following initiation of antimicrobial treatment. Expert Rev Mol Diagn 15: 349-360. Link: https://bit.ly/2xog21A
- Kaman WE, Elshout G, Bindels PJ, Mitsakakis K, Hays JP (2016) Current problems associated with the microbiological point-of-care testing of respiratory tract infections in primary care. Future Microbiol 11: 607-610. Link: https://bit.ly/2VsVk8M
- Horvat RT (2010) Review of Antibiogram Preparation and Susceptibility Testing Systems. Hosp Pharm 45: 6-9. Link: https://bit.ly/3bcOSt9
- Mitsakakis K, Kaman WE, Elshout G, Specht M, Hays JP (2018) Challenges in identifying antibiotic resistance targets for point-of-care diagnostics in general practice. Future Microbiol 13: 1157-1164. Link: https://bit.ly/2XGEOEH
- Yelin I, Snitser O, Novich G, Katz R, Tal O, et al. (2019) Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat Med 25: 1143-1152. Link: https://go.nature.com/2wFzuGy
- Cunha CB (2018) The Pharmacoeconomic Aspects of Antibiotic Stewardship Programs. Med Clin North Am 102: 937-946. Link: https://bit.ly/2RC4fDK
- Syal K, Mo M, Yu H, Iriya R, Jing W, et al. (2017) Current and emerging techniques for antibiotic susceptibility tests. Theranostics 7: 1795-1805. Link: https://bit.ly/3acGwQG
- Arlett JL, Myers EB, Roukes ML (2011) Comparative advantages of mechanical biosensors. Nat Nanotechnol 6: 203-215. Link: https://bit.ly/2VzJFoq
- Boisen A, Dohn S, Keller SS, Schmid S, Tenje M (2011) Cantilever-like micromechanical sensors. Rep Prog Phys 74: 036101. Link: https://bit.ly/2RGQK5s
- Calleja M, Kosaka PM, San Paulo Á, Tamayo J (2012) Challenges for nanomechanical sensors in biological detection. Nanoscale 4: 4925-4938. Link: https://bit.ly/3cloCwU
- Hansen KM, Thundat T (2005) Microcantilever biosensors. Methods 37: 57-64. Link: https://bit.ly/34Atrjc
- Binnig G, Quate CF, Gerber CH (1986) Atomic Force Microscope. Phys Rev Lett 56: 930-933. Link: https://bit.ly/3cn1XQu
- Krieg M, Fläschner G, Alsteens D, Gaub BM, Roos WH, et al. (2018) Atomic force microscopy-based mechanobiology. Nat Rev Phys 1: 41-57. Link: https://go.nature.com/3eqWnyr
- Müller DJ, Helenius J, Alsteens D, Dufrêne YF (2009) Force probing surfaces of living cells to molecular resolution. Nat Chem Biol 5: 383-390. Link: https://go.nature.com/3bcK6f0
- Dufrêne YF (2002) Atomic Force Microscopy, a Powerful Tool in Microbiology. J Bacteriol 184: 5205-5213. Link: https://bit.ly/2xxaJwz
- Dufrêne YF (2014) Atomic Force Microscopy in Microbiology: New Structural and Functional Insights into the Microbial Cell Surface. MBio 5: e01363-14. Link: https://bit.ly/3bfVbfq
- Formosa-Dague C, Duval RE, Dague E (2018) Cell biology of microbes and pharmacology of antimicrobial drugs explored by Atomic Force Microscopy. Semin Cell Dev Biol 73: 165-176. Link: https://bit.ly/3b6vBcJ
- Odermatt PD, Hannebelle MTM, Eskandarian HA, Nievergelt AP, et al. (2020) Overlapping and essential roles for molecular and mechanical mechanisms in mycobacterial cell division. Nat Phys 16: 57-62. Link: https://go.nature.com/2RC6EhK
- Alsteens D, Dufrêne YF (2017) Biophysics: Rapid mass changes measured in cells. Nature 550: 465-466. Link: https://bit.ly/2Rzle9F
- Etayash H, Khan MF, Kaur K, Thundat T (2016) Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat Commun 7: 12947. Link: https://go.nature.com/2VbEQD1
- Wu S, Liu X, Zhou X, Liang XM, Gao D, et al. (2016) Quantification of cell viability and rapid screening anti-cancer drug utilizing nanomechanical fluctuation. Biosens. Bioelectron 77: 164-173. Link: https://bit.ly/2VxbVrQ
- Longo G, Rio LM, Trampuz A, Dietler G, Bizzini A, et al. (2013) Antibiotic-induced modifications of the stiffness of bacterial membranes. J Microbiol Methods 93: 80-84. Link: https://bit.ly/2Vw12GL
- Stupar P, Opota O, Longo G, Prod'hom G, Dietler G, et al. (2017) Nanomechanical sensor applied to blood culture pellets: a fast approach to determine the antibiotic susceptibility against agents of bloodstream infections. Clin Microbiol Infect 23: 400-405. Link: https://bit.ly/3eq9Gz5
- Kohler AC, Venturelli L, Kannan A, Sanglard D, Dietler G, et al. (2020) Yeast Nanometric Scale Oscillations Highlights Fibronectin Induced Changes in C. albicans. Fermentation 6: 28. Link: https://bit.ly/3bdOv1l
- Kasas S, Ruggeri FS, Benadiba C, Maillard C, Stupar P, et al. (2015) Detecting nanoscale vibrations as signature of life. Proc Natl Acad Sci 112: 378-381. Link: https://bit.ly/2K9adHQ
- Lissandrello C, Inci F, Francom M, Paul MR, Demirci U, et al. (2014) Nanomechanical motion of Escherichia coli adhered to a surface. Appl Phys Lett 105: 113701. Link: https://bit.ly/2XEZcpI
- Mustazzolu A, Venturelli L, Dinarelli S, Brown K, Floto RA, et al. (2019) A Rapid Unraveling of the Activity and Antibiotic Susceptibility of Mycobacteria. Antimicrob Agents Chemother 63 pii: e02194-18. Link: https://bit.ly/2z0wHIN
- Villalba MI, Stupar P, Chomicki W, Bertacchi M, Dietler G, et al. (2018) Nanomotion Detection Method for Testing Antibiotic Resistance and Susceptibility of Slow-Growing Bacteria. Small 14. Link: https://bit.ly/34AA9FS
- Sackmann EK, Fulton AL, Beebe DJ (2014) The present and future role of microfluidics in biomedical research. Nature 507: 181-189. Link: https://bit.ly/2XBoDIJ
- Guan A, Hamilton P, Wang Y, Gorbet M, Li Z, et al. (2017) Medical devices on chips. Nat Biomed Eng 1: 0045. Link: https://go.nature.com/2RDSLj2
- Busche JF, Möller S, Stehr M, Dietzel A (2019) Cross-Flow Filtration of Escherichia coli at a Nanofluidic Gap for Fast Immobilization and Antibiotic Susceptibility Testing. Micromachines 10. Link: https://bit.ly/2V8mgvw
- Lee WB, Chien CC, You HL, Kuo FC, Lee MS, et al. (2019) An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip 19: 2699-2708. Link: https://bit.ly/3adT62a
- Pitruzzello G, Thorpe S, Johnson S, Evans A, Gadêlha H, et al. (2019) Multiparameter antibiotic resistance detection based on hydrodynamic trapping of individual E coli. Lab Chip 19: 1417-1426. Link: https://rsc.li/3ciLsF7
- Baller MK, Lang HP, Fritz J, Gerber C, Gimzewsk JK, et al. (2000) A cantilever array-based artificial nose. Ultramicroscopy 82: 1-9. Link: https://bit.ly/3bavwoh
- Barton RA, Ilic B, Verbridge SS, Cipriany BR, Parpia JM, et al. (2010) Fabrication of a Nanomechanical Mass Sensor Containing a Nanofluidic Channel. Nano Lett 10: 2058-2063. Link: https://bit.ly/3eobDfx
- Dufrêne YF, Ando T, Garcia R, Alsteens D, Martinez-Martin D, et al. (2017) Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol 12: 295-307. Link: https://go.nature.com/2K7UbhG
- Longo G, Alonso-Sarduy L, Rio LM, Bizzini A, Trampuz A, et al. (2013) Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat Nanotechnol 8: 522-526. Link: https://go.nature.com/3ekuhVP
- Martínez-Martín D, Fläschner G, Gaub B, Martin S, Newton R, et al. (2017) Inertial picobalance reveals fast mass fluctuations in mammalian cells. Nature 550: 500-505. Link: https://go.nature.com/2VbfXY8
- Steffens C, Leite FL, Bueno CC, Manzoli A, Herrmann PSDP (2012) Atomic Force Microscopy as a Tool Applied to Nano/Biosensors. Sensors 12: 8278-8300. Link: https://bit.ly/2Vy3Xyz
- Tamayo J, Kosaka PM, Ruz JJ, San Paulo Á, Calleja M (2013) Biosensors based on nanomechanical systems. Chem Soc Rev 42: 1287-1311. Link: https://bit.ly/2ycg2lb
- Voumard M, Venturelli L, Borgatta M, Croxatto A, Kasas S, et al. (2020) Adaptation of Pseudomonas aeruginosa to constant sub-inhibitory concentrations of quaternary ammonium compounds. Environ Sci Water Res Technol. Link: https://bit.ly/3ep820Q

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
