Glob J Biotechnol Biomater Sci
A Protocol for the Computational Design of High Affinity Molecularly Imprinted Polymer Synthetic Receptors
Kal Karim*, Todd Cowen, Antonio Guerreiro, Elena Piletska, Michael J Whitcombe and Sergey A Piletsky
Cite this as
Karim K, Cowen T, Guerreiro A, Piletska E, Whitcombe MJ, et al. (2017) A Protocol for the Computational Design of High Affinity Molecularly Imprinted Polymer Synthetic Receptors. Glob J Biotechnol Biomater Sci 3(1): 001-007. DOI: 10.17352/gjbbs.000009Molecularly imprinted polymer (MIP) nanoparticles, commonly referred to as ‘plastic antibodies’ or synthetic receptors, are polymeric materials with strong affinity and selectivity for a particular chemical target. MIPs are regularly produced for use in sensors for monitoring food quality and environmental pollutants, and in the design of robust and affordable replacements for biological receptors, enzymes and antibodies in drug testing and assays. More recently the easy production of MIP nanoparticles has also permitted research relating to possible in vivo applications, primarily in drug delivery systems, toxin sequestration and pathogen inhibition. The strength of the interaction between the target and the polymer binding site is dependent on the particular monomers selected in synthesis of the MIP, and the relative concentrations of these in the pre-polymerization mixture. While computational approaches have been used to aid in MIP design previously, the methods adopted are often slow and simplistic, centring on observations of the template structure with a couple of functional monomers presumed to be appropriate. We present here an automated method of rapidly screening numerous functional monomers and effectively determining appropriate monomer ratios, while accounting for spatial discrimination in selection and dynamic parameters in optimization. Example are then given of effect MIP synthesis resulting from the protocol, and the benefits of this approach over competing methods are discussed.
Introduction
While computational methods have long been used in the design of synthetic receptors, a comprehensive approach has largely been unavailable for use as a standard protocol in the production of high affinity biomimetic materials. Molecularly imprinted polymer (MIP) synthetic receptors or ‘plastic antibodies’ are prepared by the formation of a cross-linked polymer in the presence of a molecular template. The self-assembly of functional monomers with complementary functional groups to those of the template results in formation of a pre-polymerization complex, which is stabilized by cross-linking during polymer formation. Appropriate selection of functional monomers with strong interaction energies with the template will favour the associated state, resulting in the formation of MIPs with high affinity and specificity for the target. In the absence of a rational approach to the design of MIPs, monomer selection is often made on the basis of previous experience or chemical intuition; in many cases methacrylic acid is used as the sole functional monomer due to its importance in the history of molecular imprinting. Laboratory-based approaches to the optimization of monomer compositions centre on combinatorial synthesis and screening [1,2], an approach limited by the large number of different polymers required to account for the many potentially suitable monomers.
With thousands of functional monomers commercially available or readily synthesized, a more rational approach to monomer selection was obviously required for further advancement of the field. The literature provides numerous examples where researchers have investigated the strength of monomer-template interactions through the use of molecular mechanics (MM) [3-6], molecular dynamics (MD) [7-9], and quantum mechanics (QM) [10,11], based molecular modelling techniques, but these are still limited by the sequential screening of each monomer manually.
We have developed a protocol utilizing a molecular mechanics/molecular dynamics approach to the automated screening of a library of candidate functional monomers for their interaction with a chosen template. For this purpose, we have used a commercial suite of software (SYBYL® from Tripos Inc.) that uses a small library of commercially available monomers. The validation of the described computational protocol as a means of rapid and reliable MIP design is provided by reference to many published examples of high-affinity MIPs for a diverse range of targets prepared according to this design strategy.
Incentives
The foundational principle of computational MIP design is that the stability of the template-monomer complex is directly related to the quality of imprinted sites created in the polymer after cross-linking. Much of the contemporary use of molecular modelling in the design of MIPs therefore is centred on the equation:
Where EC, ET and EM are the lowest calculable energies of the template-monomer complex, template and monomer respectively. Comparison of the different values of ΔE gives an indication of the relative stability of different components in the system, and thus provides an appropriate guide to the selection of an appropriate monomer [12-14], to find the most suitable template-monomer ratio [15-17], or both [18-20]. This method has become popular along with the use of QM-based (predominantly DFT) techniques in MIP design as a result of advances in hardware making greater computer power available. However, this approach is still time consuming and computationally demanding, and QM is for this reason associated with the screening of a relatively small number of monomers (typically five or fewer). There are rare exceptions to this in which 20 or more functional monomers, along with a number of cross-linkers, have been ranked against a particular template [21-23], but the time presumably invested in this does not lend to this being an appropriate general model of MIP design.
While QM has advantages in accuracy over the alternatives which makes it desirable in the comparison of different polymerization constituents, either directly via the above equation, or by frontier orbital analysis [24-26], which can be used as an indicator of kinetic stability [27], MM and MD have the power to perform hundreds of tasks simultaneously, such as the simulation of pre-polymerization systems consisting of thousands of molecules, and provide an analysis of interactions occurring between molecules over many nanoseconds [28]. These models are also continually being revisited, often with the adoption of techniques that are new to MIP design, such as analysis by determination of the radial distribution functions (RDFs) of atoms likely to be involved in hydrogen bond formation. RDF methods provide the distances between atoms and allows selection of appropriate chemical components in the polymerization system (monomers, solvents, etc.), when used as a tool in predicting the likelihood of successful template complexation and polymer synthesis [29-31].
The protocol described herein began development in 2000 [32], and an early form of the procedure was employed for the first time to design MIPs for creatinine [33], ephedrine [34], and microcystin-LR [35]. Dozens of papers have since been published describing the use of this protocol for a broad range of templates, with the technique being continually modified to provide a reliable method of designing high affinity imprinted polymers. Here can be seen the incremental advancements describing how molecular modelling techniques can be used to rapidly screen large databases of functional monomers in order to identify suitable candidate monomers for MIP preparation. The computational time and resources required for performing these MM and MD calculations of monomer-template interactions are modest and can produce results within a few hours. The method represents a generic procedure for the selection of monomer mixtures for the imprinting of virtually any template.
Experimental
All calculations and procedures were carried out on a desktop PC running RHEL 3.0 or later (Linux platform), executing the software package SYBYL 7.3 (Tripos Inc.). The protocol described was developed using the SYBYL software but can be adapted for application in other programs. Standard procedures are followed regarding preparation of the selected template, which may be either the whole molecule (as is typical in smaller structures) or an appropriate epitope may be used to represent the binding region of a biochemical macromolecule. These structures are often obtained from online sources such as PubChem [36], ZINC [37,38], or RCSB PDB [39], when possible to ensure the correct appropriate template geometries are presented in screening.
Automatic monomer screening
Templates constructed manually may be minimized and processed by simulated annealing using any available force field, but for greater compatibility with the LeapFrog protocol the Tripos force field and Gasteiger-Hückel charges are preferred. All structures must be available in a mol2 file format.
The monomer library can be constructed by a number of approaches. Using the SYBYL software a large number of molecules can be saved under one file name or retained in one folder easily, facilitating the writing of a script which sequentially loads a monomer, records the total internal energy of the monomer and template in isolation, forms a complex by energy minimization, and records the energy of the new arrangement before restarting with a new monomer. This process can be easily automated using a simple algorithm written in SYBYL Programming Language (SPL), and can be easily adapted for use in other software. Here however we emphasize the benefits of adapting LeapFrog for use in the screening process; Leapfrog includes a function to add an observed structure to a database (‘add fragments’), or a large number of monomer can be automatically added with simple SPL algorithms (An example script is given in Appendix 1).
Once the library is established the screening can be initiated by launching the Leapfrog program. Using the ‘dream’ mode allows greater freedom to modify parameters and ensuring the ‘calculate’ option is enabled and set to ‘all atoms’ allows observation of the whole template as opposed to the binding cavity of a macromolecule. In the ‘tradeoff’ between quality and variety the former must be maximized in the ‘tradeoff’ dialogue, including removal of ‘protected’ atoms and exclusion of desolvation and chemical synthesis effects on the binding energy. Inclusion of hydrogen bond energy must also be requested via the tailored ‘energy startup’ options.
For effective adaption of Leapfrog the ‘relative move frequencies’ must also be modified from their default settings. This largely involves simplifying the relatively sophisticated program, which builds idealized receptor binding compounds from libraries of simple synthon equivalents. By removing the option to form bonds between these simple structures in the cavity (i.e. around the template) an effective screening protocol can be developed. The frequencies of all actions are therefore set as zero, except for ‘new’, ‘twist’, ‘save’ and ‘weed’, which are typically set at 10, 5, 5 and 1 respectively.
Stoichiometric refinement by molecular dynamics
On completion of the Leapfrog run a table is produced listing each of the monomers by their binding energy to the template. The highest ranking monomer or monomers can be observed in their highest affinity positions around the template and selected appropriately for stoichiometric refinement by molecular dynamics simulation. SYBYL provides a number of methods of solvating the template in the chosen monomer(s), and a number of approaches may be taken. Here we apply the XFIT algorithm, which sequentially adds solvents around the template in a close packing arrangement. This continues until the edges of the simulation environment, a small cube of dimensions automatically determined by the dimensions of the template and monomer. Molecular dynamics simulations are performed with an NTP ensemble at 300 K for 1 ns with a dielectric constant of 1. The Tripos force field is typically used with Gasteiger-Hückel charges and a non-bonding interaction limit of 8 Å is applied. The initial velocity is set from the relevant Maxwell-Boltzmann distribution. On completion the system is minimized and the interactions between the template and the solvent monomer are observed, the complex present being indicative of the appropriate ratios for the highest affinity ratio of monomers for the polymerization mixture.
Results and Discussion
Application of the method
The protocol will be of interest to researchers involved in the design and synthesis of MIPs in any format (e.g. micro- and nano- particles, films or monoliths), and suitable for the design of high affinity MIPs for diverse templates including clinical targets (drugs), environmental/food targets (e.g. toxins) and for MIPs to be used in extreme environments. This protocol is particularly suitable for use with low molar mass templates and where the development of high affinity MIPs is required: such as (i) in the separation and purification of high-value products; (ii) analytical sample pre-treatment and solid-phase extraction; (iii) drug or fragrance release matrices; (iv) adsorbents for clinical or environmental applications; (v) sensors and assays for environmental analysis, food analysis and clinical diagnostics. The benefits for end-users of this technology have been identified within clinical analysis, in diagnostics, in pharmaceutical manufacturing and by environmental agencies.
The protocol has many advantages over other modelling techniques for monomer selection and MIP design. A library of 20 polymerizable monomers (27 accounting for charged and neutral forms) can be rapidly screened with a template in around 30 minutes, or approximately 60 minutes with a database of 100 monomers. The tasks described here can be accomplished using an unmodified desktop PC in reasonable run times, obviating the need for supercomputing facilities. In the case of rationally-designed polymers (RDPs, non-imprinted polymers bearing functional monomers selected on the basis of their interaction with the intended target), the same screening protocol can be used without the need for MD analysis, further reducing the time required for effective design. For some applications (such as in environmental and food analysis) RDPs may be preferred over MIPs as they possess a high binding capacity and reduced cost, while retaining good selectivity and affinity for the target.
Database design and automated screening
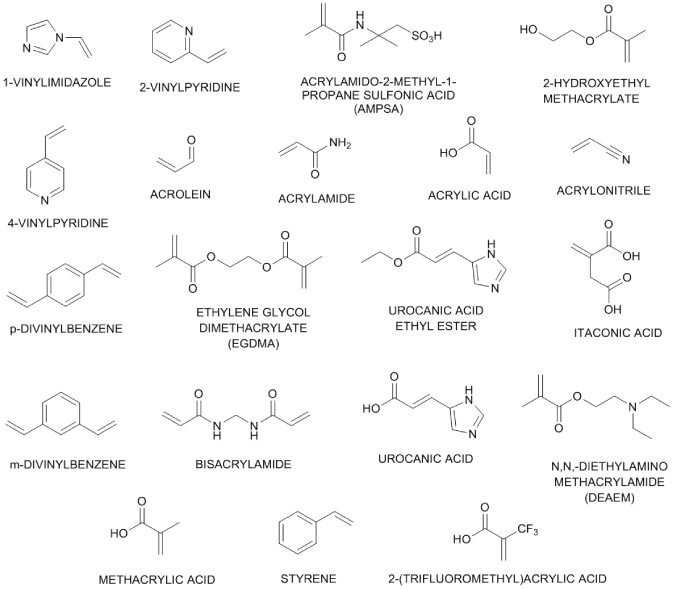
The monomer library in figure 1 is the original selection used in the automated MIP design procedure, and contains a range of acidic, basic and neutral monomers that may be capable of interacting with the template through non-covalent interactions. In this article the screening of members of the virtual monomer library for their interactions with the template is carried out using the LeapFrog algorithm within SYBYL. Existing tools for automatically ranking the greatest interaction of each of a library of compounds with another compound are not known to the authors, but the design or adaption of existing programs is possible. LeapFrog is a generic algorithm that provides a means for docking ligand precursor molecules with receptor binding sites in order to determine the optimum ligand structure. Interaction points within the receptor site are identified by sampling the environment within the binding site and determining the electrostatic, steric, and lipophilic characteristics, giving an indication of appropriate geometric and electronic properties of new drugs for the receptor. In choosing the correct parameters for the docking, the ‘ligands’ may be the library of functional monomers, and the ‘receptor sites’ regions of high or low electron density immediately surrounding the template.
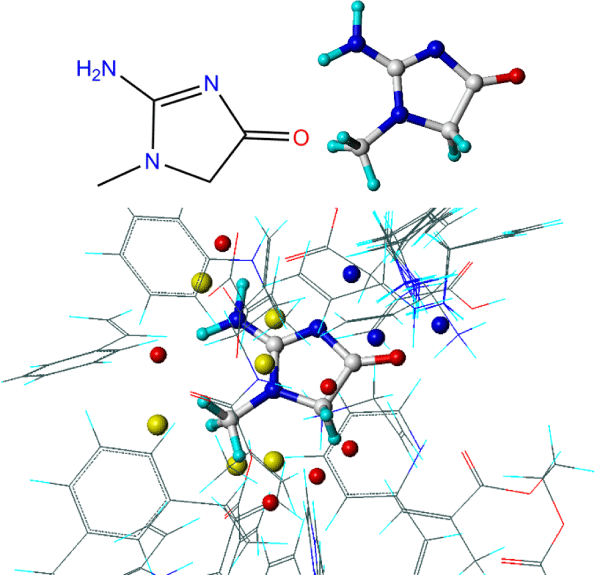
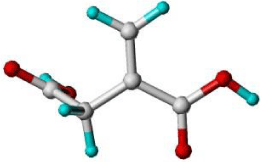
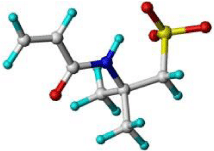
Using the LeapFrog approach, each monomer is placed in close proximity to the interaction points identified on the template surface and a binding energy calculated. The monomers are then rotated by a set small distance around the interaction point or moved to a different site, and the binding energy is again recorded. Figure 2 shows the interaction points (represented here as red, blue and yellow spheres) around a creatinine template with various monomers being analyzed. Upon completion the monomers are presented with each of their strongest binding interactions and ranked accordingly. The positions of each of the monomers, both in the position of greatest affinity and lesser arrangements, can also be visualized with the template to provide a sense of regionality. Table 1 shows a typical binding energy table ranking monomers with highest binding with N-3-oxo-dodecanoyl-L-homoserine lactone template.
Monomers giving the highest binding scores will be those that form the strongest complexes with the template and represent the best candidates for polymer preparation. By visualizing the interactions for several high affinity monomers, and each of these monomers in positions of slightly lower affinity, an indication is given of possible combinations of monomers which could be used to develop higher affinity MIPs.
Refinement using MD simulations
The appropriate ratio of polymerization mixture components is determined by performing MD simulations. This may involve the use of a single monomer species, or if the screening shows that two different monomers interact with different regions of the template, then these monomers may act synergistically in the imprinting of that template and both will then be used.
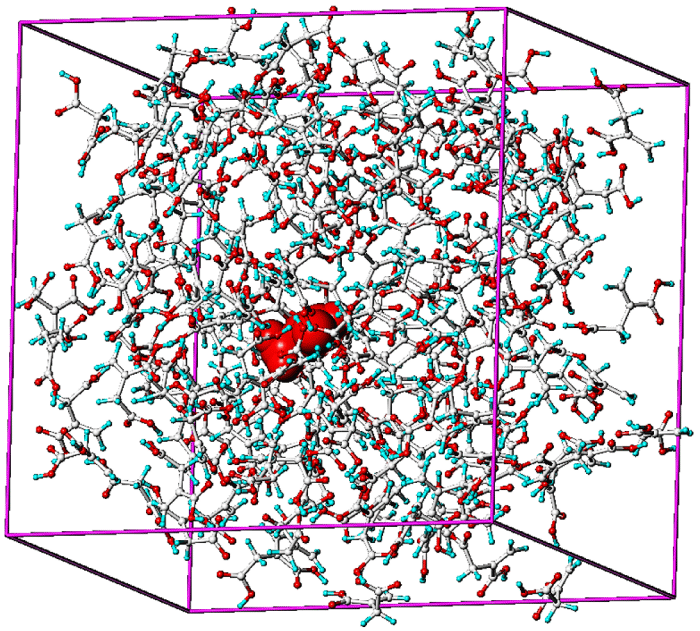
A pre-computed box of fixed dimensions is prepared by saturating the space around the template with the monomer selected from the results obtained during the screening. Figure 3 shows a graphical representation of a pre-computed box with the template at the center of the box (shown in purple) surrounded by itaconic acid. Upon equilibration the system is energetically minimized to clearly show the interactions found between the template and the monomers. Analysis of this complex provides a guide to the appropriate ratios of monomer to template in the pre-polymerization mixture. While the screening process yields good predictions of appropriate functional monomer selection alone, this process of solvating the template in the highest affinity monomer/monomers adds an additional refinement step which in not accounted for by any other approach to design.
Protocol development
The procedure was first demonstrated in a simple form some time ago for the design of a MIP for creatinine [34]. Since that time dozens of papers have been published describing the successful design of MIPs for a broad range of templates [40,41], some of which are listed in table 2 [33-35, 42-52]. The protocol has been advanced through its initial use by refinement of the parameters and the introduction of further functional monomers. A number of these new additions are more specialist compounds that must first be synthesized and cannot be readily obtained commercially, but are useful in controlling target affinity and selection for certain polymer properties.
In the case of RDPs the choice of monomer selected via the screening process has been shown to be sufficient for the synthesis of high affinity materials [43,44]. For the synthesis of these materials further refinement is not required, and so the whole design procedure can be completed in under an hour. Typically however a stoichiometric ratio must be determined for effective complexation in the pre-polymerization mixture for an imprinted polymer, and thus the MD protocol must be followed. The examples in table 2 range from (i) good imprinting factors [33,35]; (ii) high recovery of template from using solid phase extraction (SPE) [42,43,50,51]; (iii) controlled release [46,47]; (iv) dissociation constant in nM [35]; and (v) industrial applications [44,48]. This procedure therefore can be observed to produce excellent results with minimal time requirements, making this whole process highly efficient in comparison with alternative approaches.
This research used the SPECTRE High Performance Computing Facility at the University of Leicester. We would like to thank Dr Andrew Myers for his IT support.
Author Contributions
K. K., E. P. and S. P. developed the protocols. A. G. and M. W. contributed the data. K. K., T. C. and S. P. wrote the paper. All authors have discussed the results and approved the final manuscript.
- Takeuchi T, Fakuma D, Matsui J (1999) Combinatorial Molecular Imprinting: An Approach to Synthetic Polymer Receptors. Anal Chem 71: 285-290. Link: https://goo.gl/kL62Rw
- Lanza F, Sellergren B (1999) Method for synthesis and screening of large groups of molecularly imprinted polymers. Anal Chem 71: 2092-2096. Link: https://goo.gl/5axtTv
- Farrington K, Magner E, Regan F (2006) Predicting the performance of molecularly imprinted polymers: Selective extraction of caffeine by molecularly imprinted solid phase extraction. Anal Chim Acta 566: 60-68. Link: https://goo.gl/rFa3gr
- Yoshida M, Hatate Y, Uezu K, Goto M, Furusaki S (2000) Chiral-recognition polymer prepared by surface molecular imprinting technique. Colloids Surf A 169: 259-269. Link: https://goo.gl/4TmQYD
- Toorisaka E, Uezua K, Goto M, Furusaki S (2003) A molecularly imprinted polymer that shows enzymatic activity Biochem Eng J 14: 85-91. Link: https://goo.gl/i2yLsH
- Farrington K, Regan F (2007) Investigation of the nature of MIP recognition: The development and characterisation of a MIP for Ibuprofen. Biosens Bioelectron22: 1138-1146. Link: https://goo.gl/n47ygs
- Takeuchi T, Dobashi A, Kimura K (2000) Molecular Imprinting of Biotin Derivatives and Its Application to Competitive Binding Assay Using Nonisotopic Labeled Ligands. Anal Chem 72: 2418-2422. Link: https://goo.gl/SXU6Ea
- Henthorn DB, Peppas NA (2007) Molecular Simulations of Recognitive Behavior of Molecularly Imprinted Intelligent Polymeric Networks. Ind Eng ChemRes 46: 6084-6091. Link: https://goo.gl/JExbde
- Nicholls A, Andersson HS, Charlton C, Henschel H, Karlsson BCG, et al. (2009) Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens Bioelectron 25: 543-552. Link: https://goo.gl/5m65iQ
- Dong W, Yan M, Zhang M, Liu Z, Li Y (2005) A computational and experimental investigation of the interaction between the template molecule and the functional monomer used in the molecularly imprinted polymer.Anal Chim Acta542: 186-192. Link: https://goo.gl/z3Bb3d
- Ogawa T, Hoshina K, Haginaka J, Honda C, Tanimoto T, et al. (2005) Screening of bitterness-suppressing agents for quinine: The use of molecularly imprinted polymers. J Pharm Sci94: 353-362. Link: https://goo.gl/ojvwvP
- Prasad BB, Singh R, Kumar A (2016) Development of imprinted polyneutral red/electrochemically reduced graphene oxide composite for ultra-trace sensing of 6-thioguanine. Carbon 102: 86-96. Link: https://goo.gl/E9KOPw
- Tadi KK, Motghare RV, Ganesh V (2014) Electrochemical Detection of Sulfanilamide Using Pencil Graphite Electrode Based on Molecular Imprinting Technology. Electroanalysis 26: 2328-2336. Link: https://goo.gl/jTFr4H
- Qi P, Wang X, Wang X, Zhang H, Xu H, et al. (2014) Computer-assisted design and synthesis of molecularly imprinted polymers for the simultaneous determination of six carbamate pesticides from environmental water. J Sep Sci 37: 2955-2965. Link: https://goo.gl/KYbydF
- Li X, He YF, Zhao F, Zhang WY, Ye ZL (2015) Molecularly imprinted polymer-based sensors for atrazine detection by electropolymerization of o-phenylenediamine. RSC Adv 5: 56534-56540. Link: https://goo.gl/uUYu29
- Liu JB, Wang Y, Su TT, Li B, Tang SS, et al.(2015) Theoretical and experimental studies on the performances of barbital-imprinted systems. J Sep Sci 38: 4105-4110. Link: https://goo.gl/iajvP7
- Motghare RV, Tadi KK, Dhawale P, Deotare S, Kawadkar AK, et al. (2015) Voltammetric Determination of Uric Acid Based on Molecularly Imprinted Polymer Modified Carbon Paste Electrode. Electroanal 27: 825-832. Link: https://goo.gl/Nk3Zev
- Ayankojo G, Tretjakov A, Reut J, Boroznjak R, Öpik A, et al. (2015) Molecularly Imprinted Polymer Integrated with a Surface Acoustic Wave Technique for Detection of Sulfamethizole. Anal Chem 88: 1476-1484. Link: https://goo.gl/ovWcjJ
- Yang W, Liu L, Ni X, Zhou W, Huang W, et al. (2016) Computer-aided design and synthesis of magnetic molecularly imprinted polymers with high selectivity for the removal of phenol from water. J Sep Sci 39: 503-517. Link: https://goo.gl/4WAJVO
- Bujak R, Gadzała-Kopciuch R, Nowaczyk A, Raczak-Gutknecht J, Kordalewska M, et al. (2016) Selective determination of cocaine and its metabolite benzoylecgonine in environmental samples by newly developed sorbent materials. Talanta 146: 401-409. Link: https://goo.gl/O5L8ft
- Xiaoping L, Li C, Duan Y, Zhang H, Zhang D, et al. (2015) Molecularly imprinted polymer prepared by Pickering emulsion polymerization for removal of acephate residues from contaminated waters. J Appl Polym Sci 133: 43126. Link: https://goo.gl/JDa1LC
- Khan MS, Pal S, Krupadam RJ (2015) Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. J Mol Recogn 28: 427-437. Link: https://goo.gl/LWMK3J
- Wong A, Foguel MV, Khan S, Midori de Oliveira F, Tarley CRT, et al. (2015) Development of an Electrochemical Sensor Modified with Mwcnt-Cooh and Mip for Detection of Diuron. Electrochim Acta 182: 122-130. Link: https://goo.gl/200UtX
- Wang Y, Liu JB, Tang SS, Jin RF, Chang HB (2015) Preparation of Melamine Molecular Imprinted Polymer by Computer Aided Design. Chem J Chin Univ Chin 36: 945-954. Link: https://goo.gl/puCHte
- Perez M, Concur R, Ornelas M, Cordeiro MN, Azenha M, et al. (2016) Measurement artifacts identified in the UV–vis spectroscopic study of adduct formation within the context of molecular imprinting of naproxen. Spectrochem Acta Pt A-Molec Biomol Spectrosc 153: 661-668. Link: https://goo.gl/TR0TiR
- Dai ZQ, Liu J, Tang S, Wang Y, Wang Y, et al. (2015) Optimization of enrofloxacin-imprinted polymers by computer-aided design. J Mol Model 21: 290. Link: https://goo.gl/pg3F7C
- Aihara J (1999) Reduced HOMO−LUMO Gap as an Index of Kinetic Stability for Polycyclic Aromatic Hydrocarbons. J Phys Chem A 103 7487-7495. Link: https://goo.gl/PjFd4m
- Golker K, Nicholls IA (2016) The effect of crosslinking density on molecularly imprinted polymer morphology and recognition. Eur PolymJ 75: 423-430. Link: https://goo.gl/Aodtwh
- Kong Y, Wang N, Ni X, Yu Q, Liu H, et al. (2016) Molecular dynamics simulations of molecularly imprinted polymer approaches to the preparation of selective materials to remove norfloxacin. J Appl Polym Sci 133: 42817. Link: https://goo.gl/wYJUIO
- Qiu CX, Yang WM, Zhou ZP, Yan YS,WZ Xu et al. (2015) Rational design and preparation of dibenzothiophene-targeting molecularly imprinted polymers with molecular dynamics approaches and surface-initiated activators regenerated by electron-transfer atom-transfer radical polymerization. J Appl Polym Sci 132: 42629. Link: https://goo.gl/GTWh43
- Wang YC, Wang N, Ni X, Jiang Q, Yang W, et al. (2015) A core–shell CdTe quantum dots molecularly imprinted polymer for recognizing and detecting p-nitrophenol based on computer simulation. RSC Adv 5: 73424-73433. Link: https://goo.gl/dTR7YG
- Subrahmanyam S, Piletsky SA, Piletska EV, Chen B, Karim K (2001) "Bite-and-Switch" approach using computationally designed molecularly imprinted polymers for sensing of creatinine. Biosens Bioelectron16: 631-637. Link: https://goo.gl/5Ros2m
- Piletsky SA, Karim K, Piletska EV, Day CJ, Freebairn KW, et al. (2001) Recognition of ephedrine enantiomers by molecularly imprinted polymers designed using a computational approach. Analyst 126: 1826-1830. Link: https://goo.gl/0Ipjqg
- Subrahmanyam S, Piletsky SA, Piletska EV, Chen B, Karim K, et al. (2001) ‘Bite-and-Switch’ approach using computationally designed molecularly imprinted polymers for sensing of creatinine. Biosens Bioelectron 16: 631-637. Link: https://goo.gl/f3IsL2
- Chianella M, Lotierzo SA, Piletsky IE, Tothill B, Chen K, et al. (2002) Rational Design of a Polymer Specific for Microcystin-LR Using a Computational Approach. Anal Chem 74: 1288-1293. Link: https://goo.gl/7sRCtZ
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, et al. (2016) PubChem Substance and Compound databases. Nucleic Acids Res 44: D1202-D1213. Link: https://goo.gl/BlXhmF
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: A Free Tool to Discover Chemistry for Biology. J Chem Inf Model 52: 1757–1768. Link: https://goo.gl/0MHCNx
- Irwin JJ, Shoichet BK (2005) ZINC − A Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model 45: 177-182. Link: https://goo.gl/8kIcnj
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28: 235-242. Link: https://goo.gl/XZ4Qml
- S Subrahmanyam, K Karim, SA Piletsky (2013) Designing Receptors for the Next Generation of Biosensors, ed. S. A. Piletsky, M. J. Whitcombe, Springer, Berlin-Heidelberg, 131-165.
- Piletska E, Kumire J, Sergeyeva T, Piletsky S (2013) Rational design and development of affinity adsorbents for analytical and biopharmaceutical applications. Journal of the Chinese Advanced Materials Society 1: 229-244. Link: https://goo.gl/1IdWBa
- Turner NW, Pileska EV, Karim K, Whitcombe M, Malecha M, et al. (2004) Effect of the solvent on recognition properties of molecularly imprinted polymer specific for ochratoxin A. Biosens Bioelectron 20: 1060-1067. Link: https://goo.gl/t1ofQP
- Piletska E, Karim K, Coker R, Piletsky SA (2010) Development of the custom polymeric materials specific for aflatoxin B1 and ochratoxin A for application with the ToxiQuant T1 sensor tool. J Chromatogr A 1217: 2543-2547. Link: https://goo.gl/7UELLF
- Chianella I, Karim K, Piletska EV, Preston C, Piletsky SA (2006) Computational design and synthesis of molecularly imprinted polymers with high binding capacity for pharmaceutical applications-model case: Adsorbent for abacavir. Anal Chim Acta 559: 73-78. Link: https://goo.gl/hVuDNb
- EV Piletska, M Romero-Guerra, I Chianella, K Karim, APF Turner et al. (2005) Towards the development of multisensor for drugs of abuse based on molecular imprinted polymers. Anal Chim Acta 542: 111-117. Link: https://goo.gl/7Tya9G
- Breton F, Rouillon R, Piletska EV, Karim K, Guerreiro A, et al. (2007) Virtual imprinting as a tool to design efficient MIPs for photosynthesis-inhibiting herbicides. Biosens Bioelectron 22: 1948-1954. Link: https://goo.gl/ounQ11
- Piletska EV, Turner NW, Turner APF, Piletsky SA (2005) Controlled release of the herbicide simazine from computationally designed molecularly imprinted polymers. J Control Release 108: 132-139. Link: https://goo.gl/iX7y5u
- Piletsky S, Piletska E, Karim K, G Foster, C Legge et al. (2004) Custom synthesis of molecular imprinted polymers for biotechnological application: Preparation of a polymer selective for tylosin. Anal Chim Acta 504: 123-130. Link: https://goo.gl/tGJRRP
- Piletska EV, Abd BH, Krakowiak AS, Parmer A, Pink DL, et al. (2015) Magnetic high throughput screening system for the development of nano-sized molecularly imprinted polymers for controlled delivery of curcumin. Analyst 140: 3113-3120. Link: https://goo.gl/bS9vYm
- Piletska EV, Burns R, Terry LA, Piletsky SA (2012) Application of a Molecularly Imprinted Polymer for the Extraction of Kukoamine A from Potato Peels. J Agric Food Chem 60: 95-99. Link: https://goo.gl/9TydoQ
- Piletska E, Karim K, Cutler M, Piletsky S (2013) Development of the protocol for purification of artemisinin based on combination of commercial and computationally designed adsorbents. J Sep Sci 36: 400-406. Link: https://goo.gl/5K1iTu
- Piletska EV, Stavroulakis G, Karim K, Whitcombe MJ, Chianella I, et al. (2010) Attenuation of Vibrio fischeri Quorum Sensing Using Rationally Designed Polymers. Biomacromolecules 11: 975–980. Link: https://goo.gl/zLbkYo
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
