Journal of Clinical Microbiology and Biochemical Technology
Membrane and Secretory Protein Extraction of Mycobacterium Tuberculosis and Mycobacterium Bovis Using One Dimensional Electrophoresis (SDS-PAGE)
M K Sharifi Yazdi1, Mohammad Khalifeh-Gholi2, H Choobineh3, A Hadizadeh Tasbiti4*, S Sharifi Yazdi5 and Sh Yari6
2Assistant Professor, Microbiology and Immunology Department, Faculty of Medicine, Cellular and Molecular Research Center, Qom University of Medical Sciences, Qom, Iran
3Assistant Professor, Reproductive Biology, School of Allied Medical Sciences, Tehran, Iran
4Tuberculosis Department, Pasteur Institute of Iran Tehran, Iran
5Medical Student, School of Medicine, Qom University of Medical Sciences, Qom, Iran
6Tuberculosis Department, Pasteur Institute of Iran Tehran, Iran
Cite this as
Sharifi Yazdi MK, Khalifeh-Gholi M, Choobineh H, Tasbiti AH, Yazdi SS, et al. (2017) Membrane and Secretory Protein Extraction of Mycobacterium Tuberculosis and Mycobacterium Bovis Using One Dimensional Electrophoresis (SDS-PAGE). J Clin Microbiol Biochem Technol 3(2): 040-044. DOI: 10.17352/jcmbt.000025Background & Aim: Despite the drug resistance M.bovis and Mycobacterium tuberculosis (MTB) are still regarded as two of the global health problems in the world. In the present study, a comparison was made between protein profiles of M.bovis and MTB in order to achieve effective biomarkers for diagnosis of TB. The clinical samples, sputum and gastric lavage (and the other samples) were processed by N-acetyl-L-cysteine-sodium hydroxide methods and consequently were cultured on Lowenstein–Jensen medium. Mycobacterium tuberculosis and bovis strains were distinguished according to the biochemical tests and susceptibility testing system. Colonies were grown in 7H9 medium and membrane and secretory proteins were extracted, purified by ammonium sulfate and refrigerated alcohol methods. The protein contents were measured by Bradford method. Comparison of protein bands in each strain were performed by one dimensional electrophoresis. The major discrepancy between the two strains in the banding separation membrane proteins could be observed in 45 and 60 KDa and also less than 45 and 14 KDa. The results showed that discrepancy in the proteins bands could be used as protein effective biomarker for TB diagnosis. We should use antibody against TB for further investigation for rapid TB diagnosis
Introduction
Tuberculosis is a global problem. In 2014, estimates indicate that 9.6 million new TB cases, that with early diagnosis and proper treatment, nearly all of them were preventable [1]. The world “Stop TB strategy “program for the period 2006-2015 was planned to achieve this goal [2]. Mycobacterium bovis is one of the oldest and most important zoonosis diseases as well as one of the numerous global health challenges. The mortality rate of M. bovis is more than Mycobacterium tuberculosis [3].
The complex of Mycobacterium tuberculosis (MTB), including M. Tuberculosis, M. africanum, M. bovis, Bovis BCG, M. caprae, M. microti, M. pinnipedii, M. dassie bacillus and M. canettii, although shows different phenotype characteristics in their biochemical tests, but they have high similarity in genetic terms [4]. To differentiate complex members is essential to promote successful treatment , especially in areas where the disease is epidemic or exposure to human and animal is high [5]. The classic diagnosis method between M. Tuberculosis and M. bovis based on drug sensitivity, pyrazinamida activity, nitrate reduction, niacin accumulation and growth in the thiophene 2-carboxylic acid hydrazide [6]. The only way to protect people against tuberculosis are early diagnosis, protection and treatment and the current differential methods used despite their value do not meet the need for rapid diagnostic methods. Furthermore M. Tuberculosis strains resistant to antibiotics, lack of a comprehensive performance BCG vaccine in adult makes necessity for the development of rapid diagnostics and prevention will become a is a global necessity [7].One of the suitable ways in the field is the study of protein profiles of M.bovis and MTB in order to achieve effective biomarkers for diagnosis of TB. As mentioned above MTB complex shows different phenotypic profile and drug sensitive strains of M. Tuberculosis are spread almost all parts of the country that makes important from epidemiological point to study. Membrane and secretory protein have effective role in stimulating cellular immunity and are important in diagnosis of TB .M. Tuberculosis cultures filters contain various antigens in which many of them are identifiable by monoclonal antibody and are evaluated. Many of these antibodies are secreted proteins released by the cell wall or dead bacteria. These are able to generate an immune response in the early stages of infection in patients [8]. Although complete information of genomic and proteomic of M. Tuberculosis had been obtained but unfortunately not appropriate candidates for vaccine nor have protein profilie model to distinction between species been introduced.
Ying Xiong et al., had studied membrane proteins of H37Rv strains of M. Tuberculosis by one-dimensional gel electrophoresis and mass spectrometry they could integral 349 membrane protein, in which 42of them was reported and discussed for the first time [9]. The aim of this study was to evaluate differences in protein profiles of strains of Mycobacterium tuberculosis and M. bovis in order to obtained suitable biomarker to diagnosis between the two strains.
Material and Methods
In total 100 samples including 80% sputum sample,7% gastric lavage ,5% axillary node aspirate, 3.5% biopsies, 2% hip, and 1.5% bronchial washings were collected. Two standard strains of RB1P11 and 7.121 were used as control. The samples were cultivated by N-acetyl-L-cysteine in Lowensstein - Jensen Medium and thiophene2 carboxylic acid hydrazide medium were used for differentiation of M. bovis and M. Tuberculosis. After 8 weeks, Niacin, Catalase and Nitrate reduction and antibiotic susceptibility tests were carried out to identify the strains. Out of 100 tested samples, five were positive .The biochemical and drug sensitivity test showed that these strains were sensitive Mycobacterium tuberculosis. These strains were cultured in Middlebrook 7H9 medium for 4 weeks. The supernatants of stationary-phase liquid cultures were collected and used to extract proteins [10]. In order to extract membrane and secretory proteins, strains were cultured into Middle Brook 7H9 broth at 37 ° C and maintained for 4 weeks in order to get in logarithmic growth phase .After that it was centrifuged at 3000 rpm at 3°C for 45 minutes, then the supernatant was removed. The supernatant was washed with phosphate-buffered saline (PBS).The extraction buffer was (10 mM Tris containing protease inhibitors (PMSF) 1mM, DNase 1 mg and Triton x114 - 5%.The resulting solution was centrifuged at 5000 rpm at -3 °C for an hour. Composition with saturated solution of ammoniumsulfate 70% was used to distinct available proteins in the resulted supernatant and kept in Refrigerator for 18 hours. Samples were centrifuged at -4°C at 15000 rpm and obtained deposits were solved in 1×PBS.
Amount of protein in deposited solutions was obtained by Bradford protein analysis. Dialysis in 1x PBS was used to remove existing salts in the protein solutions. Protein solution inside the dialysis bag was placed in a solution of PBS 1x for 24 hours and the buffer was replaced on a regular basis, at least 3 times [11-13]. Ultimately, deposited proteins were investigated by SDS-PAGE, Blue Silver Staining and Coomasie Blue R-250. SDS- PAGE for membrane and secretory proteins was performed with 10% and 12.5 % Gel.
Results
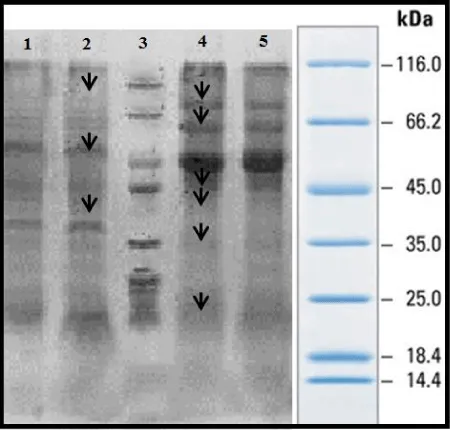
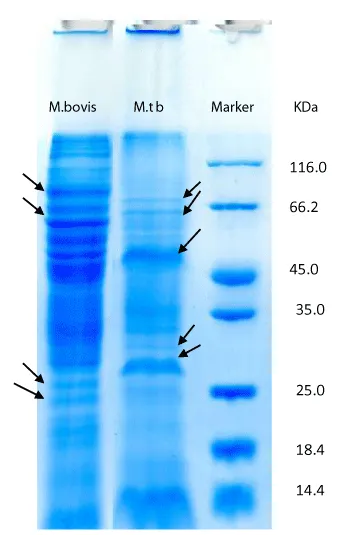
From 100 random samples cultivated in Lowenstein – Jensen solid Medium, 5 samples of susceptible M. Tuberculosis were selected by biochemical and antibiotic susceptibility testing methods along with 5 M. bovis strains cultivated in Middllebrook 7H9 medium. The concentration of proteins was defined by Bradford method after they were extracted and standard curve was drawn with bovine serum albumin 1 mg/ml as standard protein. The concentration of membrane and secretory proteins were reported 40-70 and 1-5, respectively. SDS-PAGE of membrane proteins by Gel 10% was carried out with Blue Silver Staining. It was resulted that all strains of M. bovis in 5 different patients have same protein patterns and all susceptible strains of M. Tuberculosis in 5 different patients also have same protein pattern. 15-85KDa and 15-120KDa bands were observed in strains of M. bovis and M. Tuberculosis, respectively. Differences of candidate strains of the susceptible M. Tuberculosis and M. bovis are revealed in picture (2) with arrow and it can be seen that major differences of these two strains is related to the weighted ranges of 45 and 60 KDa bands.
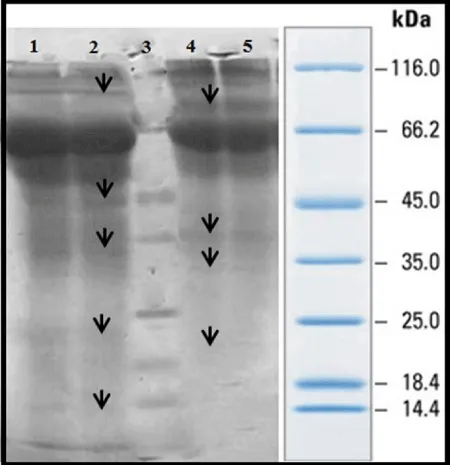
SDS-PAGE of secretory proteins were done using 12.5% Gel with Blue Silver Staining and it was perceived that all strains of M. bovis in 5 patients are with same protein pattern and all strains of susceptible M. Tuberculosis in 5 different patients also are with same pattern.
As it is shown from the 25 KDa to less than 66.2KDa Mycobacterium bovis, Mycobacterium tuberculosis strains clearly had different bands. For example, from the 45 to 50 kDa protein bands of M. bovis are differing from M. Tuberculosis. Between band 10 to 66.2 KDa only 1 or 2 bands are seen with mycobacterium tuberculosis, while 7 different protein bands are seen with Mycobacterium bovis. Results of gel electrophoresis, SDS-PAGE on 10% (Figure 1), with Blue Silver staining for membrane proteins of M. bovis and M. Tuberculosis were not same and shows identical and distinct differences in the band 14 to 25 kDa. Mycobacterium tuberculosis has some bands which is not seen in Mycobacterium bovis (protein band of 15KDa). Furthermore in region 25 to 35 KDa Mycobacterium tuberculosis had 32 to 34 KDA which cannot be seen in Mycobacterium bovis.30 kDa protein was seen in M. bovis and was not seen Mycobacterium tuberculosis. Mycobacterium tuberculosis has a 45 kDa protein that is expressed in a very clear and distinct and was not seen in M. bovi s.The50-48 kDa protein band seen M.bovis and band of around 60, 70 and above 116 kDa in Mycobacterium tuberculosis gel electrophoresis of proteins secreted by 12% and Blue Silver staining (G250) differences in protein expression between the two strains of Mycobacterium bovis strains seen. These differences in the protein bands in the area 13,23,32,44 115 kDa are seen in mycobacterium tuberculosis and not in M.bovis. On the other hand proteins with molecular weight 18,30,36,85 kDa are seen in Mycobacterium tuberculosis and not in M.bovis. Figure 2- 12.5% Gel, columns 1-5 , secretory proteins of M. bovis strains - 6-11 columns, secretory proteins of susceptible M. Tuberculosis- column 11, the marker.
In 15-115 KDa and 18-114 KDa bands were observed in strains of M. bovis and M. Tuberculosis. Differences of candidate strains are displayed by arrow in figure 3. Weighted range of 14- 45 bands is the most significant differences of these two strains.
Discussion
Considering that M. Tuberculosis diagnostic methods including susceptible and resistant to drug susceptible mycobacterium and even M. bovis are based on microbiologic methods such as direct smear, cultivation and PCR methods. But cultivation always is considered as gold standard method. The necessity of rapid treatment and easy diagnosis are inevitable considering the Low susceptibility of some of these methods such as direct view slide or long response time to suspected patients to tuberculosis including cultivation in specific mycobacterium medium. In this study we tried to compare patterns ofmembrane and secretory proteins between candidate strains of M. Tuberculosis and M. bovis using SDS- PAGE method.
The liquid Middlebrook 7H9 medium was used in the current study and makes bacteria able to synchronize in addition to making possible the study of secretory proteins. Having mineral salts, enrichment materials (such as Albumin), Catalase, dextrose, Sodium Chloride, Glycerol and Polysorbate80 are among its other advantages [14].
In 1998, Cole et al., determined the complete genome sequence of M. Tuberculosis H37Rv and defined the sequences of 3924 Gene. With the help of these genetic information, proteome analysis was done through combination of two-dimensional electrophorese and mass spectrometry [15]. Almost 800 coding secretory proteins have been recognized by M. Tuberculosis genome.
In 2003, Jens Mattow et al., analyzed supernatant proteins of mycobacterium tuberculosis cultivation using combination of two-dimensional gel electrophoresis and mass spectrometry and determined the N- terminal sequence. About 1250 protein pieces of M. Tuberculosis H37Rv were identified. This study showed 137 different proteins from which 42 proteins were explained as secretory proteins. Comparison of M. Tuberculosis H37Rv and weakened M. bovis BCG Ccopenhageh showed 39 specific protein pieces for M. Tuberculosis which had 27 different proteins and can be as a candidate of antigens in order to produce new vaccine [16].
Xing Xiong et al., investigated the membrane proteins of M. Tuberculosis H37Rv using one-dimensionalelectrophoresis and mass spectrometry and reported 349 integral fully membrane proteins and 42 membrane proteins was discussed for the first time.
Malen et al., compared membrane proteins of M. Tuberculosis H37Rv and H37Ra strains and examined the properties of more than 1700 proteins of both strains. Almost all identified proteins were too much similar, although strains were different in 5 or more proteins in 29 membrane or membrane associated proteins [17]. 19 Proteins and lipoproteins were the most frequent in H37Rv, while 10 proteins had the most frequency in H37Ra. 66 lipoproteins were the same in both strains, although 7 and 3 lipoproteins were just observed in H37Rv and H37Ra, respectively. Standard strains of ATCC and solid medium (7H10) were used in this study. Singhal et al. [18], examined the intra cellular proteins of M. Tuberculosis clinical isolates. Susceptible to drugM. Tuberculosis (susceptible to at least 5 first-line drugs of ST-11 EA13-IND family) and resistant to drug M. Tuberculosis (resistant to isoniazid rifampin and streptomycin from st288-CAS2 family) were selected from pulmonary disease center of JALMA (India). The liquid Sautons medium was used and bacteria were isolated in the late exponential phase (third week). Some protein were upregulate in comparison of 2DE and MS. 4 proteins were common in both groups AND 3 proteins belonged to metabolism and bacterial respiratory chain specifically. Results indicated that most proteins of upreguale /expressed were related to cellular metabolism and bacterial respiratory. Macrophage cell culture (THP-1) and cell infection with mycobacterium are also used. Overall, identified proteins contribute to bacteria compatibility with the environment and understanding the protein action is consistent with macrophage condition. Induction of their expression in in vitro may result in the interpretation of M. Tuberculosis strategy in creation of infection and increase of TB cell survival. Accurate identification of the proteins allows us to involve them in structural operation of TB and growth of mycobacterium in the environment Shi et al. [19], investigated the use of zinc in mice food for the cellular and humoral immune response with antigens ESAT-6 and CEP10 of Mycobacterium bovis BCG strains causes a drop in plasma cytokine levels. This has not caused changes in effect of immunization by these antigens at the time of vaccination.When comparing the bands of susceptible M. Tuberculosis in the current study with membrane proteins of M. Tuberculosis H37Rv in the Xiong study it can be state that clinical sample and standard strain of H37Rv are the same in terms of protein expression. Among main differences of the current study and the Xiong study, protein bands of 45 and 60 KDa can be noted.
It seems that the expression differences of protein bands of the two strains probably can be used as marker protein and even effective biomarker in differentiation of susceptible M. Tuberculosis and M. bovis from each other in the case of more comprehensive purification and using a suitable method because differentiation of strains is important in cases that their distinguishing is not completely clear. Treatment of patients with M. bovis and M. Tuberculosis is usually similar. Thus, difference of protein profile between these strains result in the early diagnosis and separation and consequently reducing health care costs. Selection of specific proteins able to show the differences of the strains can cause determination of these proteins to be as appropriate diagnostic biomarker. So that observation of differences in protein bands in the current study can be the beginning of different profile selection to determine the suitable candidate protein.
In this study, complementary studies including two-dimensional electrophoresis and mass spectrometry for protein difference in the observed weighted ranges will lead to more efficient purification of these Proteins in the future. Moreover, the protein difference can be useful to identify effective isotopes in immune responses of the host.
- (2015) Global tuberculosis Report.2015.World health organization.
- (2014) World Health Organization.Global tuberculosis control; ssurveillance planning, financing. Who Report? Geneva, Switzerland: WHO.
- Majoor CJ, Magis-Escurra C, Van Ingen J, Boeree MJ, Van Soolingen D (2011) Epidemiology of Mycobacterium bovis disease in humans, The Netherlands, 1993–2007. Emerg Infect Dis 17: 457-463. Link: https://goo.gl/QR8rje
- Somoskovi A, Dormandy J, Parsons LM, Kaswa M, SengGoh K, et al. (2007) Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: identification of a speciesspecific pncA mutation in Mycobacterium canettii” and the reliable and rapid predictor of pyrazinamide resistance. JCM 45: 595-599. Link: https://goo.gl/Qkg6vQ
- Barouni AS, Augusto CJ, Lopes MT, Zanini MS, Salas CE (2014) A pnca polymorphism to differentiate between Mycobacterium bovis and Mycobacterium tuberculosis.Mol Cell Probes 18: 167–170. Link: https://goo.gl/Lchp8b
- Monteros LE, Galan JC, Gutierrez M, Samper S, Garcia Marin JF, et al. (1998) Allele- specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovispnca sequence polymorphism. J Clin Microbiol 36: 239–242. Link: https://goo.gl/WcaxSe
- Barker LF, Brennan MJ, Rosenstein PK, Sadoff JC (2009) Tuberculosis vaccine research: the impact of immunology. Curr Opin Immunol 21: 331-338. Link: https://goo.gl/WrMA63
- Sonnenberg MG, Belisle JT (1997) Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N terminal amino acid sequencing, and electrospray mass spectrometry. Infect Immun 65: 4515-4524. Link: https://goo.gl/aWfVbN
- Xiong Y, Chalmers MJ, Gao FP, Cross TA, Marshall AG (2005) Identification of Mycobacterium tuberculosis H37Rv integral membrane proteins by one-dimensional gel electrophoresis and liquid chromatography electrospray ionization tandem mass spectrometry. J Proteome Res 4: 855-861. Link: https://goo.gl/ctdxQT
- Mollenkopf HJ, Grod L, Mattow J, Stein M, Knapp B, et al. (2004) Application of mycobacterial proteomics to vaccine design: improved protection by Mycobacterium bovis BCG prime-Rv3407 DNA boost vaccination against tuberculosis. Infect Immun 72: 6471–6479. Link: https://goo.gl/3Kdyfu
- kubica G, Kent P (1985) Public health microbiology A guide for the lever laboratory. Atlanta, Georgia: Public Health Service, Genters for Disease Control. Link: https://goo.gl/2pGpPd
- Farnia P, Mohammadi F, Mirsaedi M, Zia Zarifi A, Tabatabee J, et al. (2004) Bacteriological follow-up of pulmonary tuberculosis treatment: a study with a simple colorimetric assay. Microbes Infect 6: 972-976. Link: https://goo.gl/vtm3eh
- Dennison C (2002) Ed. A guide to protein isolation. New York: Kluwer Academic Publisher.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. Link: https://goo.gl/8TnpXL
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C (1998) Nature. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence 393: 537-544. Link: https://goo.gl/f5vz9a
- Mattow J, Schable UE, Schmidt F, Hagens K, Siejak F, et al. (2003) Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis 24: 3405–3420. Link: https://goo.gl/K8LiUQ
- Malen H, De Suza GA, PathakSh, Softeland T, Wiker HG (2011) Comparison of membrane proteins of Mycobacteriumtuberculosis H37Rv and H37Ra strains. BMC Microbiol. Link: https://goo.gl/jJCgXG
- Neelja S, Prashant S, Manish K, Beenu J, Deepa B (2012) Analysis of intracellular expressed proteins of Mycobacterium tuberculosis clinical isolates. Link: https://goo.gl/Bsv6WE
- Shi L, Zhang L, Li C, Hu X, Wang X, et al. (2016) Dietary zinc deficiency impairs humoral and cellular immune responses to BCG and ESAT-6/CFP-10 vaccination in offspring and adult rats 97: 86-96. Link: https://goo.gl/cgp5DH

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
