Journal of Clinical Microbiology and Biochemical Technology
Antimicrobial Resistance in Escherichia coli Isolates from Healthy Poultry, Bovine and Ovine in Tunisia: A Real Animal and Human Health Threat
Mohamed Salah Abbassi1*, Hajer Kilani1, Mohamed Zouari3, Riadh Mansouri1, Oussama El Fekih1, Salah Hammami1 and Noureddine Ben Chehida1
2University of Tunis El Manar, Faculty of Medicine of Tunis, LR99ES09 Laboratory of Antimicrobial Resistance, Tunis, Tunisia
3Office des Terres Domaniales de Mateur, Tunisia
Cite this as
Abbassi MS, Kilani H, Zouari M, Mansouri R, Oussama EF, et al. (2017) Antimicrobial Resistance in Escherichia Coli Isolates from Healthy Poultry, Bovine and Ovine in Tunisia: A Real Animal and Human Health Threat. J Clin Microbiol Biochem Technol 3(2): 019-0023. DOI: 10.17352/jcmbt.000021A total of 174 E. coli isolates collected from healthy poultry, bovine and ovine recovered between December 2009 and June 2013 in different geographic location in Tunisia, were assessed and examinated for resistance to antimicrobial agents. Avian isolates showed the highest rates of antibiotic resistance: tetracycline (74.7 %), trimethoprim/sulfamethoxazole and amoxicillin with the same rate of resistance (57 %). Prevalences of resistance to the same four antimicrobials in bovine isolates were 33.3 %, 65 %, 30 %, 28.3 %, respectively. However, for ovine isolates, low resistance rates were observed, except for tetracycline (40 %) and amoxicillin (22.85 %). Only one ESBL-producing isolate from chicken was detected. In addition, seventy-seven (44.2 %) isolates were resistant to three or more classes of antibiotics and were considered multidrug resistant (MDR). Interestingly, avian E. coli isolates were more resistant than bovine and ovine ones. These results provide novel insights into the epidemiological characteristics of poultry, bovine and ovine E.coli isolates in Tunisia, and suggest the need for the prudent use of antimicrobial agents in husbandry and the urgent need to establish a national antibiotic resistance monitoring program.
Introduction
Multi-drug-resistant bacteria have emerged as a major concern for human and veterinary medicine. To fight against this worrisome problem, reliable data on the rates of antimicrobial resistance in bacteria isolated from both human and animal are required. Therefore, it is necessary to develop monitoring programs to survey for the objective the extent of antimicrobial resistance [1,2]. Continued surveillance of the antimicrobial susceptibility profiles of foodborne pathogens, including Escherichia coli, has been strongly recommended to identify emerging antimicrobial-resistant phenotypes [3]. E. coli is a common component of the gut flora of food animals that can serve as an indicator for the acquisition of resistance to various antibiotics by enteric organisms [4,5]. The use of E. coli as the indicator bacteria is also appropriate because changes in the antibiotic resistance of this species may serve as an early warning of the development of resistance by related pathogenic bacteria [5]. Although, antimicrobial therapy is an available tool for treating clinical infections, antibiotic resistance in pathogenic bacteria from human, animal and environmental sources is recognized as a global problem in public health. Especially, the high degree of resistance to common antibiotics, the worldwide emergence of multidrug resistant phenotypes (MDR) and extended spectrum β-lactamases (ESBLs)-producing E.coli strains are of increasing concern [6,7]. A number of studies have reported that antimicrobial drugs commonly used for the treatment of human infections are misused in animals either for therapy or prevention, which has significant clinical implications for both human and veterinary medicine [8,9].
Resistant bacteria can compromise public and animal health and lead to severe economic losses in animal production [10,11].
Nowadays, the resistance to antimicrobial agents increased significantly for E. coli [12]. Initially, this resistance was described for certain antimicrobial agents such ampicillin, trimethoprime, sulphonamides and tetracycline [13]. However, since many years, large phenotypes of resistance have been observes such as resistance to all beta-lactams, aminosides, and quinolones.
Many studies on antimicrobial resistance in E. coli strains isolated from human origins in Tunisia have been published. In addition, same studies have been published, where genetic characterization of ESBL-producing E. coli isolates from animal origins and from food products of animal of origin. However, little is known about the antibiotic resistance in commensal E. coli from livestock. Indeed, it is very important to know the actual epidemiology of antibiotic susceptibility of E. coli from livestock in order to establish a guide for Tunisian veterinarians for the reasonable use of antibiotic in husbandry. For these reasons, we carried out this study to investigate antimicrobial resistance in E. coli isolates from healthy poultry, bovine and ovine during a period of 4 years and 6 months (June 2009 to December 2013).
Materials and Methods
Bacterial isolates and epidemiological background
Two hundred feces samples from various healthy animal species (bovine (n=50), ovine (50) and poultry (150)). Samples were collected from several regions in Tunisia (Siliana, Kef, Sidi Bouzid, Beja, Nabeul, Bousalem, Mateur, and Sousse) between June 2009-December 2013, Five grams of feces were incubated in Brain Heart Infusion (BHI, Becton-Dickinson) for 1 h at 37°C and then subcultured on Mac Conkey Agar (MSA, Becton-Dickinson) for 18-24h [14]. One colony from each positive sample with typical E. coli morphology was picked up randomly for further identification. Then, isolates were identified by classical bacteriological tests (Oxydase test, catalase test, Gram staining) and Api20E (Bio-Mérieux, France).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was carried out by the agar disk diffusion method on Mueller-Hinton Agar (Bio-Rad, France) plates according to recommendation of Clinical and Laboratory Standards Institute guidelines [15]. The antibiotics tested were the following: amoxicillin (25 μg), amoxicillin/clavulanic acid (20 μg + 10 μg), ticarcillin (75 μg), ticarcillin/clavulanic acid (75 μg + 10 μg), piperacillin (75 μg), cefoxitin (30 μg), ceftazidim (30 μg), cefotaxim (30 μg), imipenem (10 μg), streptomycin (10 UI), gentamicin (10 UI), kanamycin(30 UI), tobramycin (10 μg), tetracycline (30 UI), chloramphenicol (30 μg), colistin (50 μg), nalidixic acid (30 μg), ofloxacin (5 μg), ciprofloxacin (5 μg), trimethoprim/sulfamethoxazole (1,25 μg + 23,75 μg), and fosfomycin (50 μg) (Bio-Rad, France). E. coli ATCC 25922 was used as a control strain. The double-disc synergy test with cefotaxime or ceftazidime in the proximity to amoxicillin-clavulanic acid was used for the screening of Extended-Spectrum Beta-lactamases (ESBL) [16].
Statistical analysis
Statistical testing was performed using SPSS version 12.0. P < 0.05 was considered statistically significant.
Results
Identification of isolates
Amongst the 200 feces samples, 174 isolates of E. coli were collected. They were from: poultry (79), bovine (60) and ovine (35).
Antibiotic resistance rates
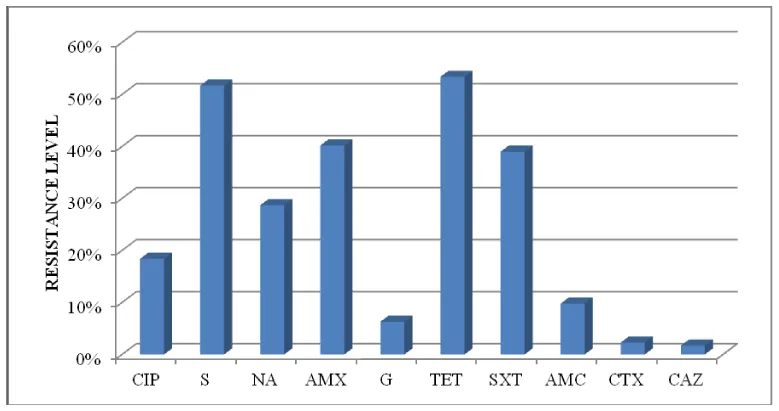
Occurrence of antibiotic resistance in the 174 E. coli isolates of is presented in table 1. High rates of resistance were observed for tetracycline (53.4 %), streptomycin (51.7 %), amoxicillin (40.2 %), and trimethoprim/sulfametoxazole (39 %). Moderate rates of resistance were observed for nalidixic acid (28.7 %) and ciprofloxacin (18.4 %). However, low frequencies of resistance were observed for amoxicillin-clavulanic acid (9.8 %), gentamicin (6.3 %), cefotaxime (2.3 %) and ceftazidime (1.7 %) (Figure 1). Interestingly, only one ESBL-producing E. coli isolate of avian origin was detected.
Analysis of resistance profile of the 174 isolates, showed that 37 isolates (21. 3 %) were susceptible to all antibiotics, 33 (19 %) were resistant to one antibiotic, 27 (15,5 %) were resistant for two antibiotics, and 77 (44,2 %) were multidrug-resistant (resistant to three or more families of antibiotics).
Rates of antibiotics resistance according to origins
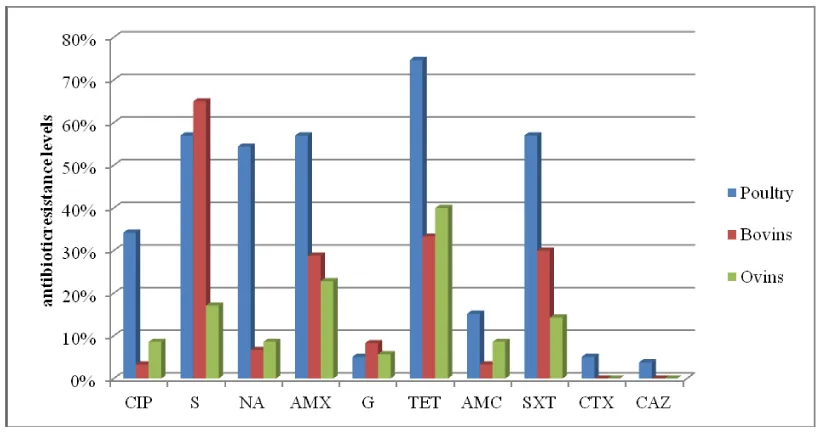
For poultry isolates, the highest rates of resistance were observed for tetracycline (74.7 %), followed by streptomycin, trimetoprime/sulfametoxazole, and amoxicillin (each 57 %), nalidixic acid (54.4 %) and ciprofloxacin (34.2 %). Whereas, much lower resistances were observed for cefotaxime, gentamicin (5.1 %), and ceftazidime (3.8 %) (Table 2). For bovine isolates, streptomycin showed an important resistance (65 %) followed by tetracycline (33.3 %), trimetoprim-sulfametoxazole (30 %), and amoxicillin (28.3 %). However, for ovine isolates, we observed low resistance rates, being, limited to tetracycline (40 %) and amoxicillin (22.85 %) (Figure 2). Taken together, we observed that avian E. coli isolates were more resistant than bovine and ovine ones, especially for tetracycline, trimethoprim/sulfametoxazole (p=10-5), amoxicillin (p=10-4), nalidixic acid (p=10-8) and ciprofloxacin (p=510-5 ).
Discussion
The aim of our study was to identify the profiles of antibiotics resistance for a collection of E. coli isolates (174) recovered from apparently healthy poultry, bovine and ovine. Our work showed high rates of resistance to tetracycline and streptomycin. Average rates of resistance were noted for amoxicillin, trimethoprim/sulfamethoxazole, nalidixic acid and ciprofloxacin. Low frequencies were observed for the resistance to amoxicilline/clavulanic-acid, gentamicin, cefotaxime and ceftazidime. A single ESBL producing strain was detected from chicken. High rates of resistance to tetracycline and streptomycin have been reported previously [17]. These antibiotics were the oldest molecules very widely used in veterinary medicine [18]. Indeed, tetracycline was usually used in the treatment of omphalites, respiratory infections, whit lows and interdigites dermatitis in ruminants as well as respiratory diseases and enteritis in poultry; it was also used as a growth promoter. Streptomycin is used for the treatment of diarrhea and streptococcal infections. These two molecules (streptomycin and tetracycline) are used very frequently and for a long time in the poultry sector. Studies made by other authors [14,19] were in agreement with our findings. Indeed, they showed a high rates of resistance to tetracycline (90.8- 95.2 %), and streptomycin (46 %). High rates of antibiotic resistance in E. coli towards tetracycline were reported at most species breeding [8,20-22]. Although frequencies of streptomycin resistance vary according to countries and conditions of breeding, the reported rates were generally very high whatever the type of animal species [8,20-22].
A moderate rate of resistance was noted for trimethoprim- sulfamethoxazole (39 %), similarly to other studies from many countries (66.1-88.6 %) [23-26]. However, a very high frequency of resistance to trimethoprim -sulfamethoxazole (80 %) was reported in our previous work [27]. Trimethoprim is used in medicine against a wide specter of bacteria including E. coli and other enterobacteria. Furthermore, the combination with sulfonamides increases its efficiency. However, the use of this antimicrobial in the animal feed, as well as its use in uncontrolled way in human medicine, during a prolonged time can entail the development and the transmission of genes encoding this resistance marker [27].
The mechanisms of resistance to beta-lactams in E. coli from animal origins is mainly encoded by extended spectrum beta-lactamases (ESBL) [28-33]. In this study, a single isolate producing ESBL from poultry origin, was identified. In Tunisia, the presence of isolates producing ESBL was reported for the first time by [34], from poultry meat, by using a selective protocol for the isolation of these isolates. However [35], did not identify ESBL-producing isolates from 164 isolates by using a non-selective protocol. [36], were the first researchers in Tunisia to report ESBL-producing E. coli from avian feces using a selective protocol. Moderate rates of resistance were observed for quinolones, nalidixic acid (28.7 %) and ciprofloxacin (18.4 %). Actually, worldwide, high rates of quinolones-resistance are increasingly reported in E. coli from animal origins. Indeed, high rates of quinolones-resistance (18.2 % - 92.5 %) were observed worldwide [14,19,37]. For ovine isolates, the frequencies of resistance were low and almost all isolates were susceptible to the tested antibiotics, except for tetracycline (40 %) and to amoxicillin (22,85 %).
Our study showed that antibiotics resistance rates in E. coli varied according to the animal species. Indeed, and for all antibiotics, except for streptomycin in bovine isolates, avian isolates were the most resistant ones, especially for tetracycline, streptomycin, trimethoprim/sulfamethoxazole, and amoxicillin. On the other hand, in cattle, the resistance rates were lower and concern only streptomycin, tetracycline and trimethoprim/sulfamethoxazole.
The results concerning the frequencies of antibiotic resistance in poultry are in agreement with other studies [14,19,38,39]. Those studies reported high rates of resistance to amoxicillin / ampicillin (25.8 %-50.4 %), tetracycline (90.8 %-95.2 %), trimethoprim-sulfamethoxazole (66.1 %-88.6 %), and streptomycin (46 %). Also, it seems that E. coli isolates from bovine and ovine origins are mainly more susceptible to antibiotics than avian isolates [40]. This might be explained to the regular use of antibiotics, in avian industries, for therapeutic or preventive purposes. This common practice mainly leads to the selection of resistant clones and, afterward, for intra- and inter-spread of these clones by fecal contamination.
The multiresistant bacteria, which accumulate resistance to several antibiotics of different families, can leads to therapeutic impasses. A simultaneous resistance to several families of antibiotics is considered as a major problem in animal and human health. Indeed, amongst our isolates, 77 (44.2 %) isolates were resistant to 3 families of antibiotics or more.
In conclusion, antibiotic susceptibility of 174 E. coli isolates collected from healthy poultry, bovine and ovine showed high frequencies of resistance to tetracycline, streptomycin, amoxicillin, and to trimethoprim-sulfamethoxazole with the presence of a single isolate producing ESBL. The highest rates of resistance were observed in poultry isolates. These results can be explained by the excessive usage of antibiotics in the industrialized poultry sector and to the practices of avian extensive breeding. However, the low rates of antibiotic resistance observed in E. coli isolates from bovine and ovine is probably linked to the scarce use of antibiotics in the Tunisian’s bovine and ovine husbandries. Taken together, antibiotic resistance in avian E. coli isolates is alarming and possible dissemination of such isolates to human and environment is worrisome.
- Allen SE, Janecko N, Pearl DL, Boerlin P, Reid Smith RJ, et al. (2013) Comparison of Escherichiacoli recoveryand antimicrobial resistance in cecal, colon,and fecal samples collected from wild house mice(Musmuscu- lus). J Wildl Dis 49: 432–436. Link: https://goo.gl/tdQPdM
- Jong DA, Thmaso V, Simjee S, Godinho K, Schiessl B, et al. (2012) Pan-European monitoring of susceptibility to human-use antimicrobial agents in entericbacteria isolated from healthy food-producing animals. J Antimicrob Chemother 67: 638–651. Link: https://goo.gl/K26iJo
- EFSA and ECDC (2012) The European Union Summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in2010. Link: https://goo.gl/uTuvaj
- Aarestrup FM, Bager F, Jensen NE, Madsen M, Meyling A, et al. (1998) Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). Acta Pathologica, Microbiologica et Immunologica Scandinavica 106: 745–770.Link: https://goo.gl/frZove
- Bogaard AE, Stobberingh EE (2000) Epidemiology of resistance to antibiotics. Links between animals and humans. International Journal of Antimicrobial Agents 14: 327–333. Link: https://goo.gl/v085rj
- Johnson JR, Kuskowski MA, Smith K, O’Bryan TT, Tatini S (2005) Antimicrobial resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis 191: 1040-1049. Link: https://goo.gl/ZJsZ90
- Teshager T, Herrero IA, Porrero MC, Garde J, Moreno MA, et al. (2000) Surveillance of antimicrobial resistance in Escherichia coli strains isolated from pigs at Spanish slaughterhouses. Int J Antimicrob Agents 15: 137-142. Link: https://goo.gl/ahGMjp
- Boerlin P, Travis R, Gyles CL, Smith RR, Janecko N, et al. (2005) Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Enivron Microbiol 71: 6753-6761. Link: https://goo.gl/dZpI6R
- Yang H, Chen S, White DG, Zhao S, McDermott P, et al. (2004) Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J Clin Microbiol 42: 3483-3489. Link: https://goo.gl/ABexxE
- Morfin OR, Tinoco Favila JC, Sader HS, Salcido GL, Perez G, et al. (2012) Resistance trends in gram negative bacteria : surveillance results from twoMexicanhospitals,2005- 2010. BMCRes Notes 5:277. Link: https://goo.gl/Cgouf8
- EFSA. (2009).Joint opinion on antimicrobial resistance (AMR) focused on zoonotic infections. Scientific opinion of the European Centre for Disease Prevention and Control; Scientific Opinion of the Panel on Biological Hazards; Opinion of the Committee for Medicinal Products for Veterinary Use; Scientific Opinion of the Scientific Committee on Emergingand Newly Identified Health Risks. EFSAJ 7: 1-78. Link: https://goo.gl/hbWxu8
- Carattoli A (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53: 2227-2238. Link: https://goo.gl/004vzH
- Gupta K, Scholes D, Stamm WE (1999) Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. J Am Med A 281: 736-738. Link: https://goo.gl/Xgl4tL
- Lei T, Tian W, He L, Huang XH, Sun YX, et al. (2010) Escherichia coli isolates from food animals, animal food products and companion animals in China. Vet Microbiol 146: 85-89. Link: https://goo.gl/GielQt
- CLSI (Clinical Laboratory Standards Institute) (2015) Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Information Supplement. M100-S25. National Committee for Clinical Laboratory Standards, Wayne PA. Link: https://goo.gl/NkcJyt
- Bradford, P. A. (2001) Extended-Spectrum -lactamases in the 21st Century: Characterization, Epidemiology, and detection of this important Resistance Threat. Clin Microbiol Rev. 14: 933-951.
- Jiang X, Tao Yu, Nan Wu, Hecheng M, Lei S (2014) Detection of qnr, aac(6’)-Ib-cr and qepA genes in Escherichia coli isolated from cooked meat products in Henan, China. Link: https://goo.gl/5Q6IsU
- Sayah RS, Kaneene JB, Johnson Y, Miller R (2005) Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic-and wild-animal fecal samples, human septage, and surface water. Appl Environment Microbiol 71: 1394-1404. Link: https://goo.gl/YXPR4d
- Jiang HX, Lü DH, Chen ZL, Wang XM, Chen JR, et al. (2009) High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Veterinary J 187: 99-103. Link: https://goo.gl/9q2UOq
- Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, et al. (2003) Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obtained over a 23 year period from pigs. Antimicrob Agents Chemother 47:3214-3221. Link: https://goo.gl/Axz5qe
- Blanco JE, Blanco M, Mora A, Blanco J (1997) Production of toxins (enterotoxins, verotoxins, and necrotoxins) and colicins by Escherichia coli strains isolated from septicemic and healthy chickens: relationship with in vivo pathogenicity J Clin Microbiol 35: 2953-2957. Link: https://goo.gl/0AosUj
- Gyles CL (2008) Antimicrobial resistance in selected bacteria from poultry. Anim Helth Res. Rev 9: 149-158. Link: https://goo.gl/H7tVvX
- Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, et al. (2003) Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coliisolates from cattle, swine and poutry. J Antimicrob Chemother 52: 489-492. Link: https://goo.gl/V5OyTg
- Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, et al. (2005) Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 55: 639-644. Link: https://goo.gl/mQfb36
- Meyer E, Lunke C, Kist M, Schwab F, Frank U (2008) Antimicrobial resistance in Escherichia coli strains isolated from food, animals and humans in Germany. Infection 36: 59-61. Link: https://goo.gl/YCDMl8
- Van Thi TH, Moutafis G, Tran LT, Coloe PJ (2007) Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl Environ Microbiol 73: 7906-7911. Link: https://goo.gl/zPWLK7
- Soufi L, Abbassi MS, Sáenz Y, Vinué L, Somalo S, et al. (2009) Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathogens and Disease 6: 1067-1073. Link: https://goo.gl/TBZCPL
- Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, et al. (2008) Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli in Belgian broiler farms. Antimicrob Agents Chemother 52: 1238-1243. Link: https://goo.gl/WEYrFy
- Smet A, Martel A, Persoons D, Dewulf J, Hevndrickx M, et al. (2009) Comparative analysis of extended-spectrum-b-lactamase (ESBL)-carrying plasmids from different members of Enterobacteriaceae isolated from poultry, pigs and humans: evidence for a shared β-lactam resistance gene pool?. J Antimicrob Chemother 63: 1286-1288. Link: https://goo.gl/XlmKCI
- Sáenz Y, Briñas L, Domínguez E, Ruiz J, Zarazaga M, et al. (2004). Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother 48: 3996-4001. Link: https://goo.gl/wza2Zu
- Brinǎs L, Moreno MA, Zarazaga M, Porrero C, Sǎenz Y, et al. (2003) Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob Agents Chemother 47: 2056-2058. Link: https://goo.gl/YqOjHn
- Li XZ, Mehrotra M, Ghimire S, Adewoye L (2007) β-Lactam resistance and β- lactamases in bacteria of animal origin. Vet Microbiol 121: 197-214. Link: https://goo.gl/OSzxi3
- Canton R, Gonzales-Alba JM, Galan RC (2012) CTX-M-enzymes: Origin and diffusion. Frontiers Microb. 3:110. Link: https://goo.gl/cJUagm
- Jouini A, Vinué L, Ben Slama K, Sáenz Y, Klibi N, et al. (2007) Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother 60: 1137-1141. Link: https://goo.gl/el0Iw3
- Soufi L, Sáenz Y, Vinué L, Abbassi MS, Ruiz E, et al. (2011) Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Inter J Food Microbiol 144: 497-502. Link: https://goo.gl/9f5fTX
- Mnif B, Ktari S, Rhimi FM, Hammami A (2012) Extensive dissemination of CTX-M-1- and CMY-2-producing Escherichia coli in poultry farms in Tunisia. Lett Appl Microbiol 55: 407-413. Link: https://goo.gl/Mw7T32
- Thorsteinsdottir TR, Haraldsson G, Fridriksdottir V, Kristinsson KG, Gunnarsson E (2009) Prevalence and genetic relatedness of antimicrobial-resistant Escherichia coli isolated from animals, foods and humans in Iceland. Zoonoses Public Health 57: 189-196. Link: https://goo.gl/3NzqSK
- Ayaz ND, Gencay YE, Erol I (2015) Phenotypic and genotypic antibiotic resistance profiles of Escherichia coli O157 from cattle and slaughterhouse wastewater isolates. Annals Microbiol 65: 1137-1144. Link: https://goo.gl/cwh7AU
- Goncuoglu M, Omranci FS, Ayaz ND, Erol I (2010) resistance of Escherichia coli O157:H7 isolated from cattle and sheep. Annals Microbiol 60: 489-494. Link: https://goo.gl/9BKVA9
- Enne VI, Cassar C, Sprigings K, Woodward MJ, Bennett PM (2007) A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol Lett 278: 193-199. Link: https://goo.gl/EXXsSL
- Int J Food Microb. Link: https://goo.gl/2dDtfH
- Pitout JD (2012) Extraintestinal pathogenic Escherichia coli: a com- bination of virulence with antibiotic resistance. Front Microbiol. Link: https://goo.gl/ji7s96
- Poirel L, Villa L, Bertini A, Pitout JD, Nordmann P, et al. (2007) Expanded-spectrum beta-lactamase and plasmid-mediated quinolone resistance. Emerg Infect Dis 13: 803-805. Link: https://goo.gl/fSML6c

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
