Journal of Clinical Microbiology and Biochemical Technology
Identification and Evaluation of In vitro Probiotic Attributes of Novel and Potential Strains of Lactic Acid Bacteria Isolated from Traditional Dairy Products of North-West Himalayas
Kanika Sharma, Nivedita Sharma* and Ranjana Sharma
Cite this as
Sharma K, Sharma N, Sharma R (2016) Identification and Evaluation of In vitro Probiotic Attributes of Novel and Potential Strains of Lactic Acid Bacteria Isolated from Traditional Dairy Products of North-West Himalayas. J Clin Microbiol Biochem Technol 2(1): 018-025. DOI: 10.17352/jcmbt.000011Background: Fermented dairy products of north-west Himalayas (Himachal Pradesh) are rich sources of potential and novel probiotic isolates which further can be explored for their commercial application.
Results: The two potential isolates Pediococcus acidilactici KM0 (accession number- KX671557) and Lactobacillus casei KL14 (KX774469) isolated from traditional fermented dairy products- noni-milk cream and lassi had shown a great potential as probiotics by exhibiting different desirable traits i.e. (that is) low pH tolerance, bile tolerance, autoaggregation, co-aggregation with pathogenic bacteria, cell surface hydrophobicity against O-xylene, antimicrobial activity against challenging food borne pathogens and antibiotic sensitivity thus advocating them as robust candidates for commercial application after going through clinical trials.
Main findings: Both the probiotic strains isolated from dairy products identified as Pediococcus acidilactici KM0 (accession number- KX671557) and Lactobacillus casei KL14 (KX774469) had been first time reported here to be potential probiotics exhibiting high acidity tolerance i.e. pH 1, 2 and 3, resisting high bile salt concentration i.e. upto 2.0%. The two isolates expressed good autoaggregation capacity i.e. greater than 40% after 5 h and showed strong hydrophobicity towards xylene i.e. > 40% with overall high cumulative score of 91.6 and 95.8% respectively.
Conclusion: The desirable attributes of these new probiotic strains strongly favor the possibility of their survival in harsh and complex habitat i.e. gastrointestinal tract (GI tract) and thus can be of commercial interest.
Brief summary: Characterization of selected lactic acid bacteria for probiotic characters (acid tolerance, bile tolerance, autoaggregation, coaggregation, hydrophobicity and antibiotic sensitivity) was done to categorize them as robust probiotics and both of these first time isolated strains had shown cumulative score as high as of 91.6 and 95.8% thus turning them highly desirable from pharmaceutical industry point of view.
Abbreviations
GI tract: Gastrointestinal tract; LAB: Lactic Acid Bacteria; WHO: World Health O; FAO: Food and Agriculture Organization; Etc: Et cetera; i.e.: That is; Viz.: That is to say; h: Hour’ w/v: Weight by volume; C: Celsius; NCBI: National Centre for Biotechnology Information; BLAST: Basic Alignment Search Tool; MTCC: Microbial Type Culture Collection; IGMC: Indira Gandhi Medical College; MRS: de Man, Rogosa and Sharpe; rpm: Revolutions Per Minute; min: Minutes; PBS: Phosphate Buffer Saline; ml: Milliliter; cfu: Colony forming unit; PUM: Phosphate Urea Magnesium Sulphate; nm: Nanometer; OD: Optical Density; M: Molar; Fig: Figure; H2O2: Hydrogen peroxide; µg: Microgram; EU: European Union; SCAN: Scientific Committee on Animal Nutrition
Introduction
The lactic acid bacteria (LAB) are generally considered as ‘food grade’ organisms and show special promise for selection and implementation as protective cultures. There are many potential applications of protective cultures in various food systems and one of that is to impart them the special status of probiotics after their rigorous characterization [1]. Probiotics are defined by World Health Organization/Food and Agriculture Organization [2] as “live microorganisms which when administered in sufficient amount confer health benefits to the host”.
Himachal Pradesh- a hilly state of north-west Himalayas is having very diverse climatic conditions that differ from semi tropical (lower hills) to semi arctic (Lahaul Spiti and Kinnaur). It is situated at an Altitude ranging from 350 meters to 6975 meters above mean sea level. The Latitude ranges from 30.2240˚ N to 33.1220˚ N whereas its Longitude ranges from 75.4555˚ E to 79.0420˚ E. In Himachal, a wide range of traditional fermented food products such as Bhaturu, Siddu, Seera, Chilra, Gulgule, Pakk, Sepubari, Bari, Aska, etc. are prepared and consumed. Among different fermented foods, fermented dairy products are the widely explored foods because of their long history of safe uses. The LAB’s isolated from dairy products are used as starter culture in the food industries i.e. fermented milk, sourdough, silage and alcoholic beverages, etc [3]. They are also added or are naturally present in raw milk and other dairy products such as yogurts, cheese and fermented milk, etc. to generate a health benefit for the consumer [4]. The staple diet of traditional fermented foods consumed by the people belonging to the tribal belts of Himachal Pradesh such as Lahaul and Spiti, Kinnaur, rural areas of Kullu, Chamba, Shimla, Kangra and Mandi form important constituents play a vital role in maintance of proper public health due to their potential enriched microflora. Fermented food products are regarded as significant part of the functional foods market and their beneficial effects are mainly associated with the maintenance of the healthy gut microbiota and an improvement of its resilience, as well as the modulation of lactose intolerance, bowel function and gastrointestinal comfort, diarrhea prevention and symptoms alleviation and regulation of immune response [5,6]. Other physiological benefits of probiotics enriched therapy include carcinogens removal, cholesterol lowering effect, immunostimulating and allergy preventing effect, synthesis and enhancing the nutrients bioavailability [7]. Probiotics are nowadays developed as novel oral vectors for mucosal delivery strategies hence act as attractive alternative to attenuated pathogens. Therefore, keeping that in mind, the present study was aimed to isolate and screen novel lactic acid bacteria from least explored traditional dairy products of Himachal Pradesh- a part of north western Himalayas and to assess their probiotic ability.
Materials and Methods
Food sources
Lassi is a fermented milk which is prepared by churning milk to create a frothy layer of butter and separating remaining residual liquor from it while milk cream-noni is fermented malai (fatty layer) of milk. The samples were collected from different districts of Himachal Pradesh and pooled individually for enrichment of culture further isolation of lactic acid bacteria. The health benefits and nutritional value of lassi and milk cream is due to the indigenous starter cultures (lactic acid bacteria) present in them.
Source of cultures
The LAB strains i.e. KM0 and KL14 were isolated from noni-milk cream and lassi. The cultures were maintained by biweekly transfers into sterile skim milk medium at 1% level by inoculating at 37oC (celsius) for 24h (hour) and held at -4°C between transfers.
Phenotypic characterization
Morphological characterization: Phenotypic characters viz. (that is to say) colour, form, margin, elevation and texture of selected isolates were taken into account after their growth of MRS (de Man, Rogosa and Sharpe) media.
Physiological characterization
(i) Growth at different temperatures
Effect of temperature on viability of probiotic isolates was examined by inoculating them in MRS broth and incubating at 15oC, 37oC and 50oC. After the incubation period of 24 h, growth was visually recorded based on intensity of turbidity.
(ii) Growth at different salt condition
Overnight grown active cultures were inoculated in MRS broth tubes adjusted to various concentrations of NaCl (sodium chloride) viz. 2, 4 and 6.5% (weight /volume, w/v) along with their controls. The cultures were incubated at 37oC .After the incubation period of 24h, growth was visually recorded based on intensity of turbidity.
(iii) Growth at different pH
To check growth at various pH, MRS broth with different pH 4.5, 6.5 and 6.5 was prepared, active cultures were inoculated in MRS broth and then incubated at 37oC for 24 h. After incubation period, extent of growth was recorded based n visible turbidity.
Genotypic identification
The sequence analysis of 16S ribosomal RNA (ribonucleic acid) gene technique (16S rRNA) was employed for identification of isolates KM0 and KL14. Then the sequence homologies were analysed by comparative studies using “The National Centre for Biotechnology Information” (NCBI) using web link (https://www.ncbi.nlm.nih.gov/) and Basic Alignment Search Tool (BLAST).
Antimicrobial activity
Challenging food borne pathogens/spoilage causing bacteria viz. Staphylococcus aureus IGMC (Indira Gandhi Medical College), Enterococcus faecalis MTCC (Microbial Type Culture Collection) 2729, Listeria monocytogens MTCC 839, Clostridium perfringens MTCC 1739, Leucononstoc mesenteroids MTCC 107 and Bacillus cereus IGMC were used as test strains to study antagonistic potential of the isolates by well diffusion assay by Kimura et al. (1998) [8].
Tolerance to low pH
The tolerance of the strains to simulated gastric juices was tested as described by Gotcheva et al. (2002) [9]. The isolates were grown on MRS broth and incubated for 24 h at 37oC. The cells were harvested by centrifugation at 5,000 rpm (revolution per minute) for 10 min (minutes) at 4oC, washed twice in sterile phosphate buffer saline PBS (phosphate buffer saline), the cells were resuspended in PBS by lowering pH to 1, 2 and 3 followed by incubation at 37oC in each set of pH for 30, 60 and 90 min. Total viable count was determined before and after incubation period at corresponding pH.
Tolerance to bile salt concentrations
The resistance of the strains to bile salt was performed according to Dora and Glenn (2002) [10]. The cells of each isolate (109 cells/ml) were inoculated separately into sterilized 10 ml MRS broth (i.e. 1% concentration) containing 0.3, 0.7, 1.0, 1.5 and 2% bile salt and incubated at 37oC for 24 h. 1ml (milliliter) of culture was taken out from each tube immediately at 0, 1, 2 and 3 h time intervals and dilutions were prepared in sterile 9 ml of 0.1% peptone water. Appropriate dilutions were pour plated in MRS agar and incubated at 37°C for 24 h and viable cells counts were noted and expressed in colony forming units (colony forming unit, cfu/ml).
Cell surface hydrophobicity
Bacterial adhesion to hydrocarbons was determined by the method of Mishra and Prasad (2005) [11]. The cells of each strain were harvested separately after growing for 18h at 37oC followed by centrifugation for 15 min at 5000 rpm. The cells were washed twice in Phosphate Urea Magnesium Sulphate (PUM) buffer and suspended separately in the same buffer at the level of 108cfu/ml. The absorbance of the suspension was measured at 600 nm (nanometer) (A). Five ml of cell suspension was mixed with 1 ml of different hydrocarbon viz. xylene, toluene, ethyl acetate and chloroform. The mixture was vortexed for 1 min and the phases were allowed to separate for 1 h at 37oC. The lower aqueous phase was carefully removed with a sterile Pasteur pipette and final absorbance (A0) was recorded at 600 nm to calculate cell hydrophobicity:
Cell auto-aggregation
The autoaggregation capacity of KM0 and KL14 was determined according to Kos et al. (2003) [12]. The cultures were grown separately in MRS broth for 18 h at 37oC. The cell suspension was centrifuged at 10,000 rpm at 4oC for 10 min. Pellet was collected and washed twice in sterile phosphate buffer saline (PBS). Cells were re-suspended in PBS, mixed by gentle vortexing for 10s and optical density (OD) was set to 0.5 at 600 nm followed by incubation at 35oC for 5 h. Absorbance of upper suspension was measured after each hour. An aliquot 0.1ml of upper suspension was taken and 3.9 ml of PBS was added to it.
Autoaggregation % was measured as 1- (At/A0) × 100, where At represents the absorbance at time t=1, 2, 3, 4, 5 h and A0 the absorbance at t = 0 h (i.e. 0.5).
Cell coaggregation
Co-aggregation ability of each isolate was determined by following the method described by Del Re et al. (2000) [13] with minor modifications. Each isolate was inoculated into MRS broth and food borne pathogens used as test indicators i.e. L. monocytogens, C. perfringens and B. cereus were inoculated in nutrient broth followed by incubation at 35˚C for 24 h. Bacterial suspension of isolates and indicators were diluted to OD=1 (λ =600nm). Bacterial suspensions were centrifuged at 10,000 rpm for 10 min at 4˚C and the cells obtained were washed three times with 0.1 M (molar) phosphate buffer saline followed by re suspension in the same buffer. Combinations of each lactic acid bacteria with all three test indicators were then made in the ratio of 1:1 to study their coaggregation ability. Each individual isolate and indicator bacteria were kept as control and were incubated at 35˚C for 4h. Absorbance at λ =600 nm was observed for mixture and each of individual strain.
Where x and y represent pathogen & LAB in the control tubes and (x + y) the mixture.
Antibiotic susceptibility test
Pattern of resistance/susceptibility to antibiotics of selected isolates KM0 and KL14 were studied by disc diffusion method as recommended by Clinical and Laboratory Standards Institute [14]. A total of 13 antibiotic discs (HiMedia Ltd, Mumbai, India) i.e. Tetracycline (TE), Cefotoxime (CTX), Amikacin (AK), Gentamycin (GEN) , Ciprofloxacin (CIP) , Ofloxacin (OF), Amphotericin-B (AP), Vanomycin (VA), Ampicillin (AMP), Erythromycin (E), Clindamycin (CD), Penicillin-G (P) and Co-trimaxazole were used. Diameter (mm) of zone of inhibition was measured using antibiotic zone scale and results were expressed in terms of resistance or susceptibility by comparing with the interpret zone diameters given by Performance Standards for Antimicrobial Disk Susceptibility tests.
Results and Discussion
Phenotypic characterization
As Himachal Pradesh is a rich repository of indigenous dairy products. Traditional dairy products have been least studied and these are also supposed to be treasure hunts of novel microorganisms, therefore isolation of lactic acid bacteria KMO and KL14 (Plates 1,2) was carried out with rarely explored fermented dairy products i.e. lassi and noni-milk cream. The morphological characters i.e. color, form, elevation, margins and texture of bacterial isolates showed that both the colonies were cream, punctiform, raised, entire and smooth. Similarily, Kumar and Kumar, (2014) [15] isolated 30 bacteria from different dairy products from Solan district in Himachal Pradesh and found out that all the colonies appeared off white, shiny white to creamy white in color, margins of these colonies were entire with flat, raised and convex elevations. The isolates were translucent and opaque in nature. Crittenden et al. (2005) [16] isolated lactic acid bacteria on MRS medium from different sources- commercial dairy food products viz. yoghurt and cheese in New Zealand. All isolates appeared as round, opaque, creamy or milky white colonies.
Morphological characterization and their tentative identification
Morphological characterization of isolated lactic acid bacteria had been done and their characteristics were noted down in Table 1. Mode of growth of isolated anaerobic lactic acid bacteria was ascertained on the basis of their sensitivity to oxygen. The isolates were found out to be facultative anaerobic. Gram’s staining has been performed to check the gram’s reaction and shape of the bacteria. The gram reactions of the isolates were determined by light microscopy after gram staining. Since, LAB’s are known to be gram positive, hence, the cells of bacterial isolates appeared blue-purple in color after gram staining. The results showed that both the isolates were gram positive. Out of these 2 isolates, KM0 was found to be cocci bacteria while KL14 was rod. Further catalase test of the isolates was performed and both were found to be catalase negative. Catalase is an enzyme produced by many microorganisms that breaks down the hydrogen peroxide into water and oxygen and causes gas bubbles. The formation of gas bubbles indicates the presence of catalase enzyme. Kumar and Kumar, (2015) [17], showed that LBS 2 colonies isolated from household curd were round, smooth, raised, entire margins, gram positive rods arranged in clusters, non-spore forming and non-motile. Savedboworn et al. (2014) [18] observed total 82 isolates of lactic acid bacteria from fermented vegetables on de Man, Rogosa and Sharpe (MRS) agar. All isolates were Gram-positive, coccus, facultative anaerobic, catalase-negative and homofermentative in nature.
Genotypic identification
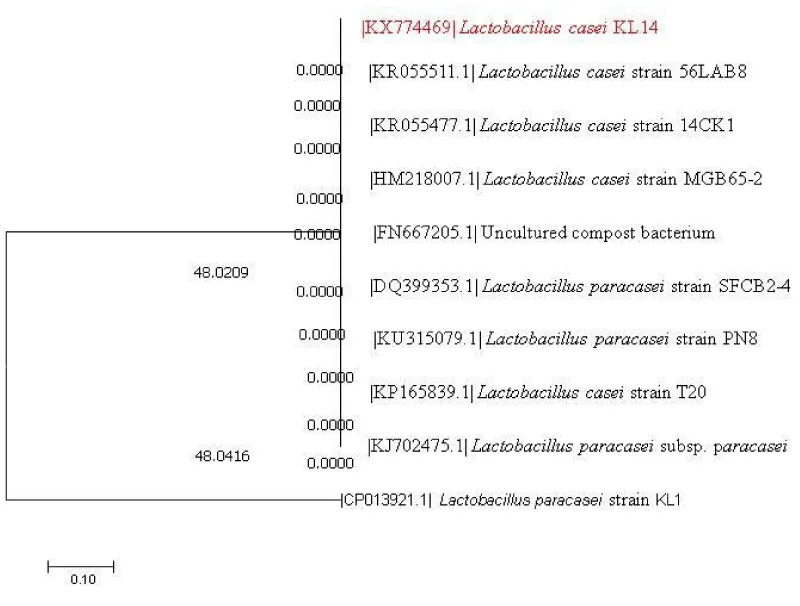
16S rRNA analysis of isolates: Sequence similarity search for the KMO (BLAST, NCBI) showed 98% homology with the available sequence of Pediococcus acidilactici accession number KX671557. Phylogenetic tree of P. acidilactici KM0 with respect to other lactic acid bacteria as inferred by neighbour joining method has been presented in Figure 1.
On the other hand, sequence similarity search for the KL14 (BLAST, NCBI) showed 98% similarity with the available sequence of Lactobacillus casei and was assigned accession no. KX774469 by NCBI. Phylogenetic tree of L. casei KL14 with respect to other lactic acid bacteria along with the boot strap values as inferred by the neighbour joining method has been presented in Figure 2.
16S rRNA is a reliable approach to identify the microorganisms by exploring their genetic sequences. Similar approach has been implied by many other authors to identify microbial isolates. Amirbozorgi et al. (2016) [19] isolated 2 groups of lactic acid bacteria from Iranian tradititonal dairy products and showed that according to 16S rRNA they found out to be Lactobacillus lactis helveticus and Lactobacillus brevis. Mesgari and Hosseinzadeh (2016) [20] isolated six different lactic acid bacteria from different types of traditional cheeses and identified by 16S rRNA as Lactobacillus rhamnosus, L. acidophilus, L. reuteri, L. fermentum, L. delbrueckii and Bifidobacterium lactis.
Antimicrobial activity
The agar well diffusion method was used to study antimicrobial activities of P. acidilactici KM0 and L. casei KL14 against pathogenic microorganisms. The zones of inhibition of indicator organisms tested were ranging from 16.80 to 24 mm and 13.0 to 19.0 mm in diameter as shown in Table 2. The strong antimicrobial activity of probiotic against challenging food borne pathogens viz. S. aureus, E. faecalis, L. monocytogens, C. perfringens, L. mesenteroids, B. cereus, P. syringe, P. caratovorum, E. coli and L. plantarum is a significant finding for competitively excluding or inhibiting the activities of pathogenic intestinal microflora thus making the host safer against the harmful microorganisms.
Lactobacilli eliminate pathogens by multiple activities viz. lowering pH and exhibit antagonistic effect by hydrogen peroxide production that produces a variety of antimicrobial compounds like lactic acid, acetic acid and H2O2, etc. In the present study the P. acidilactici KM0 and Lactobacillus casei KL14 culture had shown good antimicrobial activity against both gram positive and gram negative organisms. The antagonistic activity of isolated strains from traditional dairies microbiota was assessed by formation of inhibition zones against gram-positive and gram-negative pathogenic bacteria. Both two isolated strains, including L. plantarum 15HN and Enterococcus mundtii 50H displayed significant anti-pathogenic activities against indicator microorganisms [21].
Tolerance to low pH
One of the important criteria for good probiotics is the ability to tolerate high acid levels present in our stomachs. A good probiotic should withstand at least pH 3.0. During fasting the pH of stomach reaches as low as 1.5 [22]. Therefore, the effect of different pH on the viability of P. acidilactici KM0 and L. casei KL14 had been studied and depicted in Tables 3,4. Both the isolates showed good tolerance power at low acidic conditions (i.e. 1, 2 and 3 pH). A significantly very high resistance of L. casei KL14 followed by P. acidilactici KM0 is of special interest to place these lactic acid bacteria under the category of robust probiotics. In similar type of work by Kumar and Kumar, (2015) [17] showed that LBS 2 isolated from household curd survived acid tolerance upto 60.52%. Gautam and Sharma (2014) [23], it was observed that L. spicheri G2 had potential to tolerate pH 3.0 but a rapid decline in viability with reduction of 5 log count was observed at pH 1.0 after 1h.
Tolerance to high bile concentrations
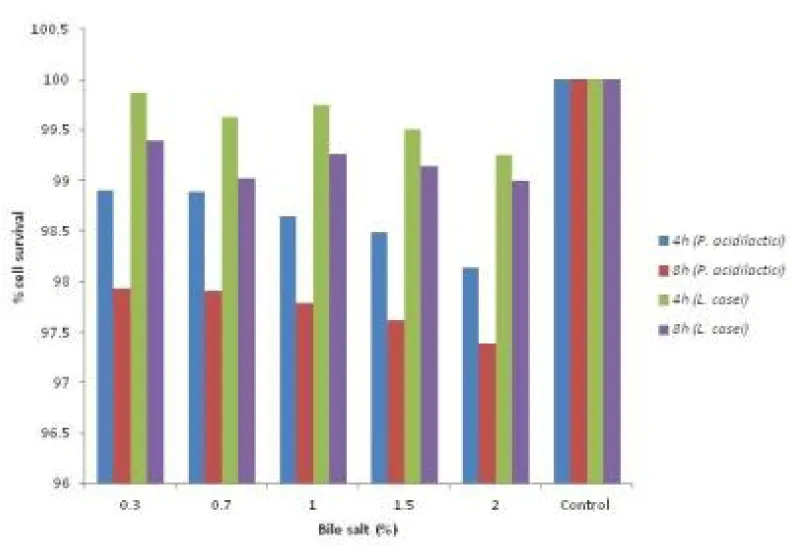
Tolerance to bile is an important feature that enables lactic acid bacteria to grow in gastrointestinal conditions. Gastrointestinal system has varying concentrations of bile ranging from 0.5% to 2.5% in the first hour of digestion and the levels may decrease further in subsequent hours [24]. Therefore, tolerance to bile salts is considered to be a prerequisite for colonization and metabolic activity of bacteria in the small intestine of the host. In the present study, it was observed that the P. acidilactici KM0 and L. casei KL14 cultures had survived and tolerated bile salts (0.3-2%) quite effectively as shown in Figure 3. In similar study, Houque et al. (2010) [25] studied Lactobacillus sp. isolated 4 isolates from yogurts and found that all the isolates were able to tolerate bile acid at the rate of 0.3%.
Bacterial adhesion to solvents (Cell hydrophobicity)
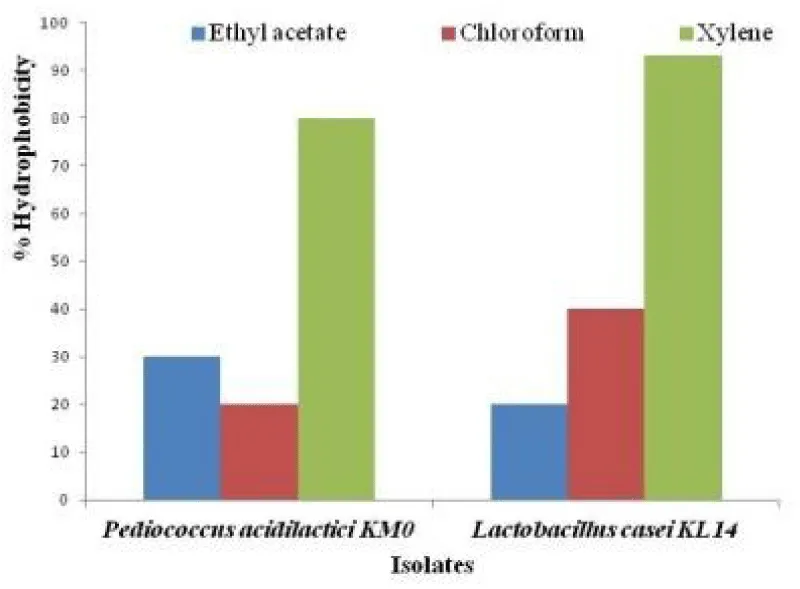
The adherence to gut is an important criterion as far as probiotic bacteria are concerned. Indeed, the probiotic ability to adhere to the intestinal epithelium is regarded as a prerequisite to colonize the human GIT (gastrointestinal tract) for exerting beneficial effects of probiotics such as the exclusion of enteropathogenic bacteria [26,27]. The ability of probiotics to adhere to epithelia is studied in vitro by evaluating the cells surface hydrophobicity toward O-xylene, chloroform and ethyl acetate. Cell surface hydrophobicity is a nonspecific interaction between microbial cells and host. The initial interaction may be weak, followed by adhesion processes that are mediated by more specific mechanisms involving cell surface proteins and lipoteichoic acids [28,29]. Bacterial adhesion to xylene, chloroform and ethyl acetate is tested to assess the Lewis acid-base characteristics of the bacterial cell surfaces. P. acidilactici KM0 showed 80% adhesion towards O-xylene, 20% adhesion towards chloroform, and 30 % towards ethyl acetate whereas L. casei KL14 exhibited 93% adhesion towards xylene, 40% adhesion towards chloroform and 20% adhesion towards ethyl acetate as shown in Figure 4. Similar results were reported by Gautam and Sharma (2014) [23], where they showed that L. spicheri G2 showed high affinity towards xylene which is a polar solvent and demonstrated hydrophobic cell surface which is a highly desirable probiotic attribute.
Autoaggregation
Autoaggregation ability determines the capacity of the bacterial strain to interact with itself in a nonspecific way. Bacterial aggregation between cells of the same strain (autoaggregation) is of considerable importance and is one of the key factors to check the ability of the lactic acid bacterial strain to adhere to the oral cavity and gastrointestinal tract, where lactic acid bacteria are to be active and provide their beneficial effects [30]. The sedimentation rate of P. acidilactici KM0 and L. casei KL14 were measured over a period of 5 h. Initially, the percentage of autoaggregation was 12.5% and 26% that increased continuously throughout the testing span and finally registered a high percentage of 50 % and 80% respectively (Table 5). These strains were found as strong autoaggregating phenotypes. The observed autoaggregation could be related to cell surface component, because this property had not been lost after washing and suspending of the cells in phosphate buffer saline. Similarily, Kabore et al. (2012) [31] measured sedimentation rate of L. helveticus B9 and L. acidophilus BS isolated from commercial yogurts over a period of 5 h and the results revealed that autoaggregation ability was high, i.e. 80.29±1.67% and 77.58±1.46%, respectively.
Co-aggregation activity
Probiotics are able to achieve coaggregation with pathogenic microorganisms and in turn effectively inhibit and kill them by secreting antimicrobial compounds that act directly on the cells of pathogenic bacteria [32]. In this study, the coaggregation ability of P. acidilactici KM0 and L. casei KL14 has been studied and their percent coaggregation with pathogens were calculated as 31.0 (L. monocytogenes), 15.0 (C. perfringens) and 5.2 (B. cereus) respectively. P. acidilactici KM0 exhibited highest coaggregation ability with pathogenic bacteria L. monocytogens (31.0%) followed by 15 and 5.2% by C. perfringens and B. cereus respectively whereas L. casei KL14 exhibited 23.0% highest coaggregation percent with L. monocytogens, 25% with B. cereus and 12.5 % with C. perfringens (Table 6). In similar study, Gupta and Sharma, (2015) Pediococcus pentosaceus C-1 showed co-aggregation with Listeria monocytogenes (18.36%), Bacillus cereus (14.2%) and Clostridium perfringens (14.0%).
Antibiotic susceptibility test
The results of antibiogram of the culture investigated using disc diffusion assay for a total number of twelve clinically important antibiotics according to Performance Standards for Antimicrobial Disk Susceptibility tests presented in Table 7. P. acidilactici KM0 and L. casei KL14 were found susceptible to majority of antibiotics namely Ampicillin (10 µg), Gentamycin (10 µg), Penicillin G (10 µg), Clindamycin (30 µg), Erythromycin (15 µg), Tetracycline (30 µg), Cifrofloxacin (5 µg), Oflaxacin (10 µg), Amphotericin B (100 µg) and Vancomycin (30 µg). The antibiotic susceptibility of strains is crucial from the safety point of view for their use as potential probiotics because antibiotic resistant probiotic bacteria may act as potential reservoirs of antimicrobial resistance genes which can be transferred to pathogenic bacteria. As per the European Union (EU) Scientific Committee on Animal Nutrition (SCAN) guidelines, bacteria used in food/feeds should not contain any acquired antibiotic resistances. In this study, P. acidilactici KM0 and L. casei KL14 were overall found to be sensitive to the most of the antibiotics thereby showing no risk of having antibiotic resistance gene. The results obtained were well in agreement with that of Handa and Sharma, (2016) [34] in which Lactobacillus plantarum F22 was found to be sensitive to majority of tested antibiotics viz. Ampicillin (10 µg), Gentamycin (10 μg), Nalidixic (30 μg), Amoxyclove (30 μg), Chloromphenicol (30 μg), Tetracycline (30 μg), Cifrofloxacin (5 μg), Co-trimoxazol (25 μg), Methicillin (30 μg), and Vancomycin (30 μg), etc. Zdolec et al. (2008) [35], who also recorded Lactobacillus sp. susceptibility to antibiotics viz. tetracycline, penicillin, bacitracin, chloramphenicol, cefalexin, linkomycin, erythromycin, ampicillin, spiramycin and amoxicillin.
Cumulative probiotic potential of probiotic lactic acid bacterial isolates
Cumulative probiotic score has been provided to these isolates based upon the desirable attributes exhibited by these isolates. Cumulative probiotic potential is the sum of score of acid tolerance, bile salt tolerance, autoaggregation, hydrophobicity, antibiotic susceptibility and antagonistic potential. The probiotic potential for screened exopolysaccharide producing bacterial isolates was calculated by following formula:
In the present study, probiotic potential of P. acidilactici KM0 and L. casei KL14 was adjudged very high i.e. 91.6 and 95.8%. Generally, commercially available probiotic preparations have probiotic score in the range of 75-85%. Since, both probiotic lactic acid bacteria in the present study have qualified the score strongly and thus these are labeled as good probiotics with high prospective for commercial applications as per criteria of FAO/WHO (2002) [2]. The present investigation has meet out the status of good probiotics.
Conclusions
Results obtained in this study showed that both probiotic strains i.e. P. acidilactici KM0 and L. casei KL14 isolated from dairy products fermented milk cream-noni and lassi had maximum acid tolerance, bile tolerance, cell autoaggregation, antimicrobial activity and were sensitive to maximum number of antibiotics used. Their high probiotic score prove them as strong candidates for commercial application with different desirable health benefits.
- Holzapfel WH, Haberer P, Geisen R, Bjorkroth J, Schillinger U (2001) Taxonomy and important features of probiotic microorganisms in food and nutrition. Am J Clin Nutr 73: 365S–373S. Link: https://goo.gl/Cm7oiv
- FAO/WHO (2002) Guidelines for the evaluation for the probiotics in food: report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. Link: https://goo.gl/oSi7kP
- Carr FJ, Chill D, Maida N (2002) The lactic acid bacteria, a literature survey. Critical Reviews in Microbiology 28: 281-370. Link: https://goo.gl/YUzfgW
- Oskar A, Meydani SN, Russell RM (2004) Yogurt and gut function. Am J Clin Nutr 80: 245-256. Link: https://goo.gl/wyRYH3
- Mohammed S, Ahlgren JA, Horne D (2016) Structural characterization and biological activities of an exopolysaccharide kefiran produced by Lactobacillus kefiranofaciens WT-2B(T). J Agri Food Chem 52: 5533-5538. Link: https://goo.gl/FV8oJN
- Ouwehand A, Salminen S, Isolauri E (2002) Probiotics: an overview of beneficial effects. A Van Leeuw 82: 279-289. Link: https://goo.gl/s1MfJ6
- Vasiee AR, Tabatabaei Yazdi F, Mortazavi A, Edalatian MR (2014) Isolation, identification and characterization of probiotic Lactobacilli sp. from Tarkhineh. Int Food Res J 21(6): 2487-2492. Link: https://goo.gl/XekIKK
- Kimura H, Sashihara T, Matsusaki H, Sonomoto K, Ishizaki A (1998) Novel bacteriocin of Pediococcus sp. ISK-1 isolated from well – aged bed of fermented rice bran. Ann N Y Acad Sci 864: 345-348. Link: https://goo.gl/NWlZhv
- Gotcheva V, Hristozova E, Hristozova T, Guo M, Roshkova Z, Angelov A (2002) Assessment of potential probiotic properties of lactic acid bacteria and yeast strains. Food Biotechnol 16: 211-225. Link: https://goo.gl/OdpxfH
- Dora IAP, Glenn RG (2002) Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl Environ Microb 68: 4689- 4693. Link: https://goo.gl/JAAY2q
- Mishra V, Prasad D (2005) Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics. Int J Food Microbiol 103: 109-115. Link: https://goo.gl/i1Q6Yc
- Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S (2003) Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J App Microbiol 94: 981–987. Link: https://goo.gl/A8k9AG
- Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31: 438-442. Link: https://goo.gl/Df3dO6
- CLSI (2007) Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing. 17th edition. Wayne, PA.
- Kumar A, Kumar D (2014) Isolation and characterization of bacteria from dairy samples of Solan in Himachal Pradesh for identification of Lactobacillus spp. Int J Pharm Sci Rev Res 25(2): 110-114. Link: https://goo.gl/7vitcq
- Crittenden R, Bird AR, Gopal P, Henriksson A, Lee YK, et al. (2005) Probiotic research in Australia, New Zealand and the asia-pacific region. Curr Pharm Design 11: 37-53. Link: https://goo.gl/GiWExs
- Kumar A, Kumar D (2015) Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobe 33: 117-123. Link: https://goo.gl/ZU3tPK
- Savedboworn W, Riansa-Ngawong W, Sinlapacharoen W, Pajakang S, Patcharajarukit B, et al. (2014) Assessment of probiotic properties in lactic acid bacteria isolated from fermented vegetables. Int J Appl Sci Technol 7: 53-65. Link: https://goo.gl/TxV8yR
- Amirbozorgi G, Samadlouie H, Shahidi SA (2016) Identification and characterization of lactic acid bacteria isolated from Iranian traditional dairy products. Int Brazilian Jiu-Jitsu Federation 2: 1-6. Link: https://goo.gl/AsWj7K
- Mesgari P, Hosseinzadeh (2016) Isolation and identification of Lactobacillus acidophilus and Bifidobacterium from different types of (traditional) cheese and study their antibacterial properties. Int J Adv Biotechnol Res 7: 275-281. Link: https://goo.gl/3nxEnm
- Haghshenas B, Haghshenas M, Nami Y, Khosroushahi AY, Abdullah N et al. (2016) Probiotic assessment of Lactobacillus plantarum 15HN and Enterococcus mundtii 50H isolated from traditional dairies microbiota. Adv Pharm Bull 6: 37-47. Link: https://goo.gl/t0JyEK
- Fernandez MF, Boris S, Barbes C (2003) Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol 94: 449-455. Link: https://goo.gl/Uggw0r
- Gautam N, Sharma N (2014) Quality attributes of novel cereal based probiotic products prepared by using food grade lactic acid bacteria. Indian J Tradit Know 13: 525-530. Link: https://goo.gl/sjAOuh
- Hati S, Shilpa V, Surajit M, Dipika Y, Vandana (2014) Evaluation of probiotic potentials of L. plantarum C6 isolated from cheese. Indian J Dairy Sci 67: 413-420. Link: https://goo.gl/FCn3JZ
- Hoque MZ, Akter F, Hossain KM, Rahman MSM, Billah MM, et al. (2010) Isolation, identification and analysis of probiotic properties of Lactobacillus Sp. from selective regional yoghurts. World J Dairy Food Sci 5: 39-46. Link: https://goo.gl/13AYvC
- Collado M, Meriluoto J, Salminen S (2007) In vitro analysis of probiotic strain combinations to inhibit pathogen adhesion to human intestinal mucus. Food Res Int 40: 629-639. Link: https://goo.gl/UUeAd7
- Licht TR, Wilcks A (2005) Conjugative gene transfer in the gastrointestinal environment. Adv Appl Microbiol 58: 77-95. Link: https://goo.gl/2ULr7c
- Rojas M, Ascencio F, Conway PL (2002) Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl Environ Microbiol 68: 2330-2336. Link: https://goo.gl/wbWQ5m
- Ross S, Jonsson H (2002) A high-molecular mass cell surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148: 433-442. Link: https://goo.gl/ypWiZB
- Nikolic M, Lopez P, Strahinic I, Suarez A, Kojic M, et al. (2012) Characterisation of the exopolysaccharide (EPS)-producing Lactobacillus paraplantarum BGCG11 and its non-EPS producing derivative strains as potential probiotics. Int J Food Microbiol 158: 155-162. Link: https://goo.gl/x47ZXe
- Kabore D, Sawadogo-Lingani H, Dicko MH, Diawara B, Jakobsen M (2012) Acid resistance, bile tolerance and antimicrobial properties of dominant lactic acid bacteria isolated from traditional “maari” baobab seeds fermented condiment. Afr J Biotechnol 11: 1197-1206. Link: https://goo.gl/F9vYuP
- Bao Y, Zhang Y, Zhang Y, Lui, Wang S (2010) Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 21: 695-701. Link: https://goo.gl/uN2hSs
- Gupta A, Sharma N (2015) In vitro characterization of probiotic properties and detection of novel compounds secreted by lactic acid bacteria isolated from kodo millet (Paspalum scrobiculatum) flour: A traditional Himalayan food. GERF Bulletin of Biosciences 6: 7-20. Link: https://goo.gl/6ho4Db
- Handa S, Sharma N (2016) In vitro study of probiotic properties of Lactobacillus plantarum F22 isolated from Chhang- A traditional fermented beverage of Himachal Pradesh, India. Journal of Genetic Engineering and Biotechnology 14(1): 91-97. Link: https://goo.gl/8ivzAC
- Zdolec N, Hadžiosmanović M, Kozačinski L, Cvrtila Z, Filipović I, et al. (2008) Microbial and physicochemical succession in fermented sausages produced with bacteriocinogenic culture of Lactobacillus sakei and semi-purified bacteriocin mesenterocin Y. Meat Sci 80: 480–487. Link: https://goo.gl/qwYEg0

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
