Open Journal of Proteomics and Genomics
Genetic variants of COVID-19 and vaccination. Is there a Correlation?
Nagwa Ali Sabri1*, Mohamed Ahmed Raslan2, Eslam Mansour Shehata3 and Sara Ahmed Raslan2
2Clinical Researcher -Drug Research Centre-Cairo -Egypt
3BSc, Clinical Research, Drug Research Centre, Egypt
Cite this as
Sabri NA, Raslan MA, Shehata EM, Raslan SA (2022) Genetic variants of COVID-19 and vaccination. Is there a Correlation? Open J Proteom Genom 7(1): 001-005. DOI: 10.17352/ojpg.000011Copyright License
© 2022 Sabri NA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: New coronavirus disease is considered one of the most widely spreading viral infections all over the world. Increased numbers of severe covid-19 cases are growing up. Gene sequencing and discovering new viral variants is an essential aspect during the pandemic. The generation of treatment-resistant viral strains and the probability of negative impact on vaccination efficacy is possible. We aimed to review the probable effect of new variant emergence on treatment and vaccination efficacy, besides, the importance of gene sequencing from published literature data till the moment.
Main body of the abstract: SARS-CoV-2 genome studies indicated that it shared 79 to 82% nucleotide similarity with SARS-CoV-1. Several gene locations in the envelope (E) structural protein c.222G>C (p. Leu74Leu) and the Membrane (M) structural protein c.213C>T (p. Tyr71Tyr) were proved to have mutations. Also, the surface (S) gene mutation c.1841A>G (p. Asp614Gly) is most relevant. The published sequences in Egypt are accounting for less than 0.2 percent of reported instances.
Short conclusion: The possibility of rapid generation of treatment-resistant viral strains is highly possible. As a consequence of genetic alterations that impart functional differences in infectivity, sub-strains might arise as a result of acquired immunity that is likely to diminish over time and become less effective against increasingly aggressive strains. Gene sequencing in Egypt requires a lot of efforts to provide a rapid discovery for new emerging variants, to avoid a possible decrease in vaccination efficacy and emergence of treatment-resistant strains.
Abbreviations
WHO: World Health Organization; PCR: Polymerase Chain Reaction; COVID-19: Coronavirus Disease 2019; MERS-CoV: Middle East Respiratory Syndrome Coronavirus; SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; SARS-CoV-1: Severe Acute Respiratory Syndrome Coronavirus 1; RT-qPCR: Quantitative Reverse Transcription PCR; (E) structural protein: Envelope structural protein; (M) structural protein: Membrane structural protein; (S) gene mutation: Surface gene mutation; SPs: Structural Proteins; ACE2: Angiotensin-Converting Enzyme 2; RNA: Ribonucleic Acid; DNA: Deoxyribonucleic Acid; RBD: Receptor-Binding Domain; IL-6: Interleukin 6; IL-10: Interleukin 10; TNF: Tumor Necrosis Factor; B-cells: Beta cells; Fc receptors: Fragment Crystallizable Region Receptors; ORF1ab gene: Overlapping Open Reading Frames that Encode Polyproteins 1ab; RdRp: RNA-dependent RNA polymerase; RSV: Respiratory Syncytial Virus
Background
A viral respiratory disease outbreak (officially known as coronavirus disease by the WHO) occurred in the Chinese city of Wuhan, at the end of 2019 [1].
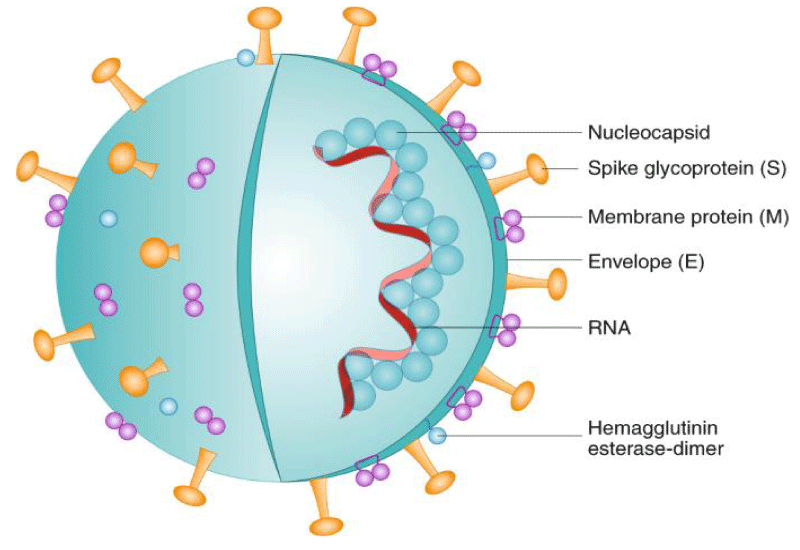
SRAS-CoV (2003) has infected 8098 people with a 9 percent fatality rate in 26 countries in the past, on the other hand, novel coronavirus (2019) infected 252,826,597 individuals with a mortality rate of 2.01%, across 109 countries, till 16th NOV 2021. It is obvious that SARS-CoV-2 has a greater transmission rate than SRAS-CoV, which might be due to a genetic recombination event at the S protein in the RBD region of SARS-CoV-2, which may have boosted its transmission potential [2,3] Figure 1.
The spread of SARS-CoV-2 or any respiratory infections can occur via three transmission modes, which are contact transmission, droplet transmission, and airborne transmission. Viral contact transmission can occur through physical contact or fomites harboring settled droplets, where, droplet transmission involves huge droplets larger than 20μm in diameter produced by a forceful expiratory incident and deposited directly on the conjunctiva or mucus membranes of a susceptible host. On the other hand, airborne transmission occurs by small respiratory droplets (aerosols) smaller than 10μm in diameter that remain airborne for a long time that is enough to transfer a pathogen and deposited deep into the respiratory tract [5].
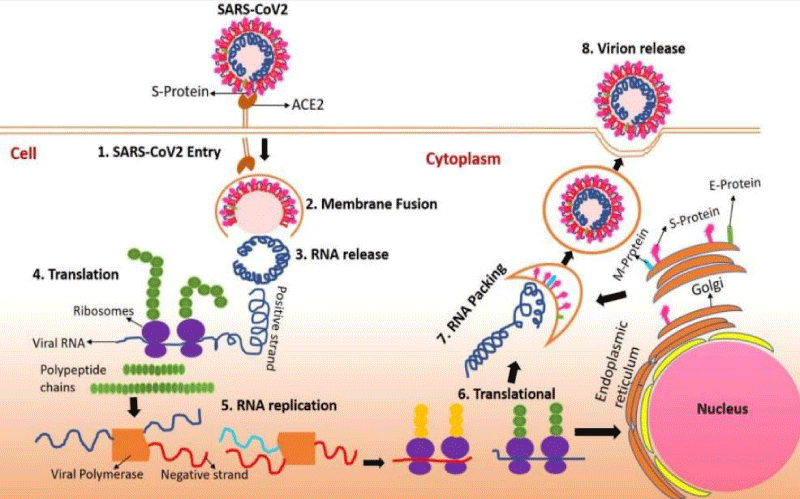
The viral entry and replication mechanism, besides RNA packing in the human cell, is attributed to coronavirus spike (S) protein attachment to Angiotensin-Converting Enzyme 2 (ACE2) receptors located on the surface of numerous human cells, particularly those in the lungs thus permitting viral entry, translation, RNA replication, and packaging, followed by virion release [6] Figure 2.
Only molecular PCR (RT-qPCR) test of respiratory tract samples is currently recommended for the identification and confirmation of COVID-19 cases since these methods have been evaluated for safety and quality through (WHO) protocols. Serological tests are used as an auxiliary approach for monitoring the epidemiological situation, despite the fact that they are faster and less expensive than genetic studies. This diagnostic approach has low sensitivity, but it is valuable for monitoring the infection, thus efforts to enhance it are continuing. Those serological tests include different types like Euroimmun SARS-COV-2 ELISA (IgG), Biomedomics COVID-19 IgM-IgG Rapid Test, Tianjin Beroni Biotechnology SARS-COV-2 IgG/IgM antibody detection kit, and others. The developed kit’s sensitivity and specificity range from 60% up to 100% [7].
Severe Acute Respiratory Syndrome Coronavirus 2 genome size ranges from 29.8 to 29.9 kb, where more than 2/3 of the genome and encoding ORF1ab polyproteins are resulting from the 50 section, while the 1/3 of genes encoding for SPs such as surface (S), envelope (E), nucleocapsid (N), and membrane (M) proteins are from the remaining 30 section [8].
The establishment of a statistical approach based on genetic knowledge besides epidemiology, and antigenic factors may give additional insight into the mechanisms governing the emergence and evolution of unique mutations, as well as increase the capability for SARS-CoV-2 prevention and management [9,10].
Coronaviruses belong to the Orthocoronavirinae subfamily, Coronaviridae family, Coronavirinae suborder, and Nidovirales order [11]. This Orthocoronavirinae subfamily is further classified into 4 genera which are alpha (α), beta (β), delta (δ), and gamma (γ) coronavirus. Until now, only six coronaviruses had been proved to cause human infections. Four of the six viruses known as human coronaviruses (HCoV) 229E, NL63, OC43, and HKU1 are infecting humans, and induce only relatively insignificant upper respiratory symptoms [12].
The remaining two viruses from the genera betacoronavirus which are (MERS-CoV) appeared in 2012 in Saudi Arabia and (SARS-CoV) developed in China during 2002 and 2003 can cause acute lower respiratory symptoms [13,14].
Gamma (γ) and delta (δ) coronaviruses, contain a variety of avian coronaviruses. According to sequencing research, SARS-CoV-2 possesses the main model of genomic make-up of coronaviruses, namely Beta coronavirus, subgenus Sarbecovirus. These coronaviruses are considered to be the 7th coronavirus to infect human beings [15].
Coronaviruses are widespread pathogens in the environment, causing gastrointestinal or respiratory illnesses and occasionally damaging essential systems in people and animals. Coronaviruses can be killed by ultraviolet radiation, elevated temperatures, 75% ethanol, ether, chloroform, and disinfectants containing chlorine [16].
The emerging COVID-19 variants not only cause elevated transmissibility, and mortality, but they also can evade detection by diagnostic tests, exhibit lowered susceptibility to treatment such as antivirals, monoclonal antibodies, and convalescent plasma. Besides, causing re-infection to previously cured and vaccinated individuals [17].
Individuals with COVID-19 who were infected with the delta form had a higher probability of hospitalization or emergency care than those who were infected with the alpha variant. According to clinical study findings, outbreaks of the delta variation among unvaccinated populations may exert a greater burden on healthcare systems than epidemics of the alpha type [18].
Early data suggests that both Omicron (B.1.1.529) and Alpha (B.1.1.7) variants have a P681H mutation, which, when combined with two other changes, may allow the virus to transmit more easily between individuals. Moreover, it was postulated that combining the Q498R and N501Y additional changes may improve the virus’s capacity to attach to the host receptor ACE2 [19].
Many nations throughout the world have prohibited or limited travel to and from South Africa, and a 14-day quarantine period has been imposed for visitors arriving from Omicron-infected areas [20].
According to reported data on COVID-19-infected patients, those who are hospitalized in intensive care units (ICUs) have concomitant comorbidities and are at a greater risk of bacterial infections. That is why there is a concern about the emergence of antimicrobial resistance as a result of high demand and misuse of antimicrobial agents in hospitals [21].
When the prevalence of COVID-19 is reduced for a while, the predicted social behavior is a reduction in or abandonment of precautionary measures on a societal level [22]. That may raise high concerns about the emergence of new viral strains that can escape the acquired vaccine immunity.
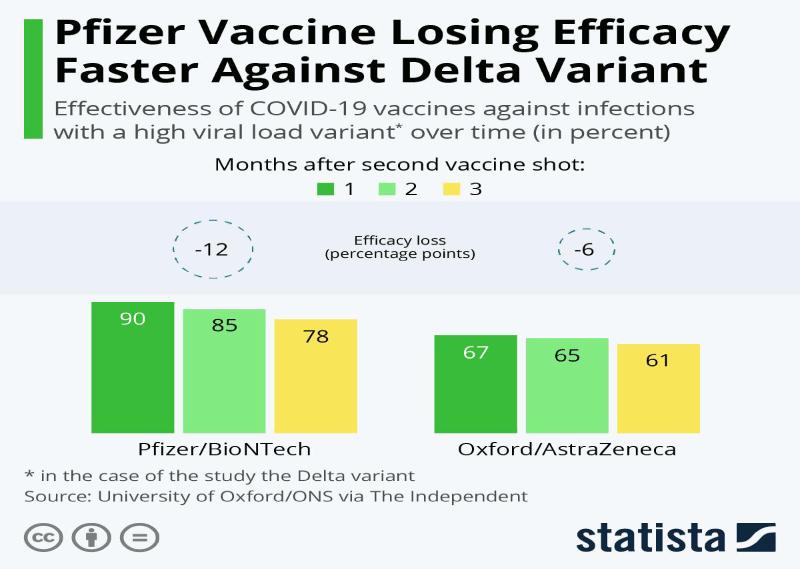
It is worthy to mention that, one issue that may impact vaccination effectiveness is the likelihood that induced immunity would decrease over time and become less effective against increasingly aggressive strains, thus, sub-strains can form as a result of genetic changes that impart functional variations in infectivity, and transmissibility [23] Figure 3.
A limited number of investigations regarding COVID-19 gene sequencing in Egypt were performed. Gene sequencing needs more support to allow discovering new variants, prevention of emergence of more resistant variants, and prevent further viral wide-spreading. The effect of genetic mutations on viral pathogenicity and vaccine efficacy will be a highlight in this review to know the current and future viral genomic variant’s effect.
Main text
Upon making a comparison between the SARS-CoV-2 genome and another human coronavirus’, studies revealed that the gene of SARS-CoV-2 is similar to human SARS-CoV-1 (nucleotide similarity of about 79 to 82 percent). On the other hand, it was determined to be far from the MERS-CoV genome [25].
In specific, SARS-CoV-2 spike protein (S1 subunit) displayed a greater binding affinity to humans (ACE-2) than that of SARS-CoV-1. The reason for such high affinity was related to the 5 amino acid residues change in the RBD of the S1 subunit that affects spike protein structure [26,27].
In a research study, synonymous changes in the c.222G>C (p.Leu74Leu) location of the E gene and c.213C>T (p.Tyr71Tyr) position of the M gene were found. A point mutation in the S and N genes has been discovered. Furthermore, the sole significant differential in surface spikes of the SARS-CoV-2 virus is the aspartate (D) mutation at position 614. This mutation was found in the majority of a group of Chinese sequences to glycine (G) which was prevalent in other Western Europe populations. Furthermore, 14 additional variants other than the D614 Gone have been found. Replication rates of the covid-19 virus can be influenced by D614 G mutation carrier strains (Orf1ab P4715L; RdRp P323L) [28,29].
These proteins are a target for drugs used as antiviral agents like favipiravir and redeliver. Besides, these proteins are prone to changes that indicate a fast generation of resistant strains. In current investigations, the most prevalent variant is the c.1841A>G (p. Asp614Gly) missense mutation which is found in the S gene. Furthermore, it is shown to have an ORF1ab P4715L replication mutation [30,31].
The D614G S-protein mutation in the carboxy(C)-terminal region of the S1 domain was previously identified to be increasing at an alarming rate. It was observed at low frequency in March by 26 percent but grew significantly by April to 65 percent and by May up to 70 percent. This behavior demonstrates the advantage in transmission versus the D614 viruses. The mutation was also linked with an elevated COVID-19 viral charge in infected patients, but S-protein involvement in these data is still unknown because this mutation is also linked with viral nsp3 and RdRp mutations [32,33].
In the Middle East and North Africa, the D614G mutation looked to be taking over COVID-19 infections. SARS-CoV-2 may have been circulating in the Middle East and North Africa earlier than previously thought, according to Bayesian research. This demonstrates the significance of careful surveillance in outbreaks caused by new viruses [34].
Egypt’s strains were classified as belonging to clade A2a. This clade also contains strains from Africa, Europe, the USA, Asia, and Australia. Quick examination in future research is needed to estimate the biological impact of genetic and amino acid variations between SARS-CoV-2 isolates from Egypt and the original Chinese strain on viral virulence, transmissibility, and current treatment approaches requires [35].
Gene sequencing efforts are relatively restricted among Egyptian research institutes, with published sequences accounting for less than 0.2% of reported instances. A national effort, organized and facilitated by a central entity, would make this type of research more routine and non-competitive. Plenty of additional logistical issues stands in the way of researchers’ ability to improve SARS-CoV-2 sequencing efforts. One of the most difficult issues is gathering geographically representative samples [36].
A genomic DNA study was performed on 70 blood samples collected from Egyptian populations, where, the results showed in exon 15 (c.1995 delT) a one frameshift mutation, in exon 16 (c.2070T>C) one identical NSP, and exon 18 NM_021804CT>GG, and exon 18 NM_021804TA>AG three splicing in ACE2 receptor. The frameshift deletions indicated here have only been detected once, and their level is insufficient to suggest protein structural alterations [37].
A typical viral characteristic is that increasing transmissibility is associated with decreasing virulence. This property is exhibited by SARS-CoV-2. This characteristic is supported by observations of infected patients in Wuhan province. At an early stage of the pandemic, patients had more severe diseases than those at the later stage. This happened also in Zhejiang province when transmission began to rise at a later period. According to SARS-CoV-2 genetic study data obtained during later phases, mutations on the surface of the spike protein (near the furin cleavage site) might alter protein shape and the binding of ACE2. This will result in the appearance of mild symptoms [38].
Different pharmaceutical firms and research centers are working on the development of coronavirus vaccines. The scope of vaccine development includes inactivated target virus, inactivated viral proteins, vector-based vaccines, and SARS-CoV-2 RNA or DNA. The target antigen is SARS-spike CoV-2’s glycoprotein or its receptor-binding domain required for viral cell entrance [39-41], different types of vaccines are available in Egypt for example Sinopharm and Sinovac vaccine (live attenuated), Astrazeneca/Oxford vaccine (Vector based).
As immunity spreads across the community by vaccination or spontaneous infection, it is likely that this pressure will lead to the selection of immune-evading mutations. This in turn will allow SARS-CoV-2 to be more endemic and frequent, but with mild severity [23].
Additionally, in antibody-dependent enhancement (ADE), non-neutralizing antibodies may promote viral entrance into cells. This can occur via Fc receptors or the complement system. This mechanism may result in the activation of B-cells, monocytes, and macrophages as well as the generation of IL-6, TNF, and IL-10 even in the absence of active viral replication in immune cells. Vaccine-induced antibody-dependent enhancement has been observed following the use of vaccinations inactivated by formalin against RSV and measles, as well as dengue virus vaccination use [42].
From the previously presented data and information, many issues have been highlighted about Antibody-Dependent Enhancement (ADE) possibility in patients with COVID-19 following immunization. The ADE is mediated by Fc receptors. Mutations in the spike glycoprotein might decrease the antibody response of the host. Besides, it could increase the likelihood of ADE. Infection of monocytes, macrophages, and B cells may occur in a variety of organs as a consequence of later unstable virus-antibody complexes. This will result in the widespread death of immune cells and the generation of cytokines [42].
Conclusion
Several genetic mutations were discovered in several gene locations. c.222G>C (p.Leu74Leu) in the E structural protein and c.213C>T(p.Tyr71Tyr) in the M structural protein were discovered. Moreover, D614 G and other additional variants have been found which might decrease the main host antibody response, The probability of fast generation of treatment-resistant strains, including antiviral resistance, is highly possible and consequently is an important raised issue concerning vaccine effectiveness and its benefits in immunization against SARS-COV-2. More support is required for facilitating and accelerating gene sequencing efforts worldwide to provide a vigilant and rapid discovery for new emerging variants.
- Pal M, Berhanu G, Desalegn C, Kandi V (2020) Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 12: e7423. Link: https://bit.ly/33adnIy
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res 24: 91-98. Link: https://bit.ly/364BUzW
- Link: https://bit.ly/3LmJ1Uy
- Florindo HF, Kleiner R, Vaskovich-Koubi D, Acúrcio RC, Carreira B, et al. (2020) Immune-mediated approaches against COVID-19. Nat Nanotechnol 15: 630–645. Link: https://bit.ly/3Bb6frW
- Priyanka, Choudhary OP, Singh I, Patra G (2020) Aerosol transmission of SARS-CoV-2: The unresolved paradox. Trav Med Infec Dis 37: 101869. Link: https://bit.ly/3LmVzew
- Subramanian B, Poma AB, Kolandaivel P (2021) Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn 39: 3409-3418. Link: https://bit.ly/34r0A5k
- Kubina R, Dziedzic A (2020) Molecular and Serological Tests for COVID-19 a Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics (Basel) 10: 434. Link: https://bit.ly/34tLJqw
- Zekri AN, Easa Amer K, Hafez MM, Hassan ZK, Ahmed OS, et al. (2021) Genomic characterization of SARS-CoV-2 in Egypt. J Adv Res 30: 123-132. Link: https://bit.ly/3Be4Fp7
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, et al. (2013) Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503: 535-538. Link: https://go.nature.com/3B9hbXf
- Yang XL, Hu B, Wang B, Wang MN, Zhang Q, et al. (2015) Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus. J Virol 90: 3253-3256. Link: https://bit.ly/3LhiJ5Y
- Gorbalenya AE, Baker S, Baric R, De-Groot RJ, Drosten C, et al. (2020) The species Severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5: 536-544. Link: https://bit.ly/34oFVih
- Park SE (2020) Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin Exp Pediatr 63: 119-124.
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814-1820. Link: https://bit.ly/3oCdfcc
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, et al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967-1976. Link: https://bit.ly/3rB4AsC
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382: 727-733. Link: https://bit.ly/3gDagvO
- Kang S, Peng W, Zhu Y, Lu S, Zhou M, et al. (2020) Recent progress in understanding 2019 novel coronavirus (SARS-CoV-2) associated with human respiratory disease: detection, mechanisms and treatment. Int J Antimicrob Agents 55: 105950. Link: https://bit.ly/3BbwkHq
- Vasireddy D, Vanaparthy R, Mohan G, Malayala SV, Atluri P, et al. (2021) Review of COVID-19 Variants and COVID-19 Vaccine Efficacy: What the Clinician Should Know? J Clin Med Res 13: 317-325. Link: https://bit.ly/34zlMpz
- Twohig KA, Nyberg T, Zaidi A, Thelwall S, Sinnathamby MA, et al. (2021) Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis 22: 35-42. Link: https://bit.ly/3oDfNal
- Dhawan M, Priyanka, Choudhary OP (2021) Omicron SARS-CoV-2 variant: Reasons of emergence and lessons learnt-Correspondence. Int J Surg 97: 106198. Link: https://bit.ly/3rF78G9
- Choudhary OP, Dhawan M, Priyanka (2022) Omicron variant (B.1.1.529) of SARS-CoV-2: Threat
assessment and plan of action. Int J Surg 97: 106187. Link: https://bit.ly/3BjS18o - Raslan M, Eslam MS, Sara AR, Nagwa AS (2020) Antimicrobial Resistance and Second Wave of COVID-19: Will It Impose Management Protocols Deviation?. Ann Pharmacol Toxicol 1: 1001. Link: https://bit.ly/3GzrTHu
- Raslan SA, Shehata EM, Raslan MA, Sabri NA (2020) Management of Second Wave of COVID 19 with Ledipasvir / Sofosbuvir Combination. Will it Work? A Review Article. Saudi Journal of Medical and Pharmaceutical Sciences 6: 654-657. Link: https://bit.ly/3B9vsmw
- Dos Santos WG (2021) Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines. Biomed Pharmacother 136: 111272. Link: https://bit.ly/3suQrwr
- Buchholz K (2021) COVID-19 Vaccines, Pfizer Losing Efficacy Faster Against Delta Variant. Statista. Link: https://bit.ly/3rGURRK
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395: 565-574. Link: https://bit.ly/3LmK5I2
- Khailany RA, Safdar M, Ozaslan M (2020) Genomic characterization of a novel SARS-CoV-2. Gene Rep 19: 100682. Link: https://bit.ly/3LoVXsX
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367: 1260-1263. Link: https://bit.ly/3JfTMpD
- Becerra-Flores M, Cardozo T (2020) SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract 74: e13525. Link: https://bit.ly/3Bj2KzN
- Koyama T, Platt D, Parida L (2020) Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 98: 495–504. Link: https://bit.ly/3Bdttxu
- Lokman SM, Rasheduzzaman M, Salauddin A, Barua R, Tanzina AY, et al. (2020) Exploring the genomic and proteomic variations of SARSCoV-2 spike glycoprotein: A computational biology approach. Infect Genet Evol 84: 104389. Link: https://bit.ly/3BapOAJ
- Laha S, Chakraborty J, Das S, Mannaet SK, Biswas S, et al. (2020) Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol 85: 104445. Link: https://bit.ly/339Q2qm
- Bal A, Destras G, Gaymard A, Bouscambert-Duchamp M, Valette M, et al. (2020) Molecular characterization of SARS-CoV-2 in the first COVID-19 cluster in France reveals an amino acid deletion in nsp2 (Asp268del). Clin Microbiol Infect 26: 960–962. Link: https://bit.ly/3rB5Qfk
- Lizhou Z, Jackson CB, Mou H, Ojha A, Rangarajan ES, et al. (2020) The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. Link: https://bit.ly/3su6aMa
- Sallam M, Ababneh NA, Dababseh D, Bakri FG, Mahafzah A (2021) Temporal increase in D614G mutation of SARS-CoV-2 in the Middle East and North Africa. Heliyon 7: e06035. Link: https://bit.ly/34ADYPA
- Kandeil A, Mostafa A, El-Shesheny R, Shehata M, Roshdy WH, et al. (2020) Coding-Complete Genome Sequences of Two SARS-CoV-2 Isolates from Egypt. Microbiology resource announcements 9: 22 e00489-20. Link: https://bit.ly/3gDvIkh
- Al-Ghazawy O (2021) Egypt’s SARS-CoV-2 sequencing challenges. Nature Middle East. Link: https://go.nature.com/3uJjcYP
- Hafez MM, Hassan ZK, Bahnasy AA, Ahmed OS, Abouelhoda M, et al. (2020) SARS-CoV-2 receptor mutation in Egyptian population. medRxiv Link: https://bit.ly/3sukeW2
- Jin X, Xu K, Jiang P, Lian J, Hao S, et al. (2020) Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg Microbes Infect 9: 1474-1488. Link: https://bit.ly/34JQkVp
- Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, et al. (2020) SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586: 567-571. Link: https://bit.ly/3uFtbyg
- Gao Q, Bao L, Mao H, Wang L, Xu K, et al. (2020) Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369: 77-81. Link: https://bit.ly/34r2bbk
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271-280.e8. Link: https://bit.ly/3oCUJAL
- Poland GA, Ovsyannikova IG, Kennedy RB (2020) SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet 396: 1595-1606. Link: https://bit.ly/3oyAvbh
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
