Open Journal of Proteomics and Genomics
3D Structure Modeling of Catalase Enzyme from Aspergillus Fumigatus
Vivek Dhar Dwivedi1*, Shiv Bharadwaj2, Aman Chandra Kaushik3 and Sarad Kumar Mishra4
2Nanotechnology Research and Application Center, Sabanci University, Istanbul, Turkey
3Schools of Biotechnology, Gautam Buddha University, Greater Noida, India
4Department of Biotechnology, D.D.U. Gorakhpur University, Gorakhpur UP, India
Cite this as
Vivek Dhar D, Shiv B, Kaushik AC, Sarad Kumar M (2016) 3D Structure Modeling of Catalase Enzyme from Aspergillus Fumigatus. Open J Proteom Genom 1(1): 008-012. DOI: 10.17352/ojpg.000002The respiratory diseases in humans, such as aspergilloma, allergic bronchopulmonary aspergillosis and invasive aspergillosis are caused by the fungal pathogen Aspergillus Fumigatus (A. fumigatus) The enzyme catalase of A. fumigatus provides a putative virulence to this fungal pathogen against the toxic effects of human hydrogen peroxide, which they cleave into water and molecular oxygen. The 3-D structure of this protein in A. fumigatus is not known, while it is very important for understanding the molecular mechanism of action of this enzyme and development of new drugs for various respiratory diseases. This article proposes the 3-D structure of catalase enzyme from A. fumigatus which has been predicted and validated using different computational programs. Based on the percentage of residue occurrence in helical, strand and loop regions, four structural domains have been identified in the modeled structure. The structure and function relationship for all identified structural domains have been also described. This study will be helpful for in silico drug discovery against the virulence nature of Aspergillus Fumigatus.
Introduction
In the last few years, several Aspergillus sp. have attracted much focus as opportunist pathogens as reflected in various published reports [1-4]. Fungi are able to cause diseases by overwhelming the host defense systems due to presence of several genes and proteins associated with their pathogenicity, called virulence factors [5]. The fungal pathogen Aspergillus Fumigatus, member of Aspergillaceae family, has been associated with a wide spectrum of diseases in human, such as allergic bronchopulmonary aspergillosis, aspergilloma and invasive aspergillosis etc. [6]. It is one of the most ubiquitous fungi due to easy dispersal of its conidia [7]. It is calculated that a person could inhale several hundred conidia of A. fumigatus per day [7]. Alveolar macrophages and polymorphonuclear cells, cellular components of innate immunity of the lung, work against the conidia and hyphal form, respectively [8]. However, in vitro studies of neutrophil function have revealed that H2O2 effectively kills fungal hyphae [9] and that neutrophil-mediated damage is blocked by addition of a commercial catalase [10]. In A. fumigatus, three active catalases have been identified in which catalase enzyme present in the conidium is encoded by cat A gene [11], and two mycelial catalases are encoded by cat1/cat B [12] and cat2 genes [11]. It was reported that A. fumigatus conidial and mycelial catalases protect the fungus against hydrogen peroxide [11]. However, the inhibition of catalase activity could make A. fumigatus vulnerable to rapid killing by the H2O2 and accordingly, catalase which is a good scavenger of H2O2, was considered to be a putative virulence factor of A. fumigatus [13]. Hence, thorough study of 3D structure of the A. fumigatus catalase enzyme present in the above fungus can be helpful in understanding the mechanism of its defense against the host immune response and also in the prediction its molecular mechanism of infection.
Materials and Methods
Protein sequence data
In the present work, the primary full-length amino acid sequences of A. fumigatus catalase enzyme was searched and retrieved from GenPept, a protein sequence database of National Centre for Biotechnology Information (NCBI) in fasta format.
3-D structure Prediction, evaluation and validation
Template selection and 3-D structure of A. fumigatus catalase enzyme was determined by HHpred server by HMM-HMM comparison [14]. The PDB file of the modeled structure was then downloaded after the completion of 3-D modeling. The model was visualized using PyMol [15]. The predicted 3-D model of A. fumigatus catalase enzyme was further validated using the programs; PROCHECK [16], ProSA [17], ProQ [18] and Profile 3-D [19]. The PROCHECK was employed to check the valid stereochemistry and ProSA to detect the native structure compatibility. Also, ProQ, a neural network-based predictor based on a number of structural features was used to calculate the quality of the generated protein 3D model. The root mean square deviation (RMSD) of each atoms of the predicted model with respect to the template structure was also calculated using SuperPose tool [19]. Besides, the overall stereochemical quality of the protein, its amino acid residues in the allowed, disallowed region and overall G-factor were also evaluated by Ramachandran plot analysis [20].
Structural annotation and domain identification
The structural domains in the modeled structure of the catalase protein were recognized using DIAL server (https://caps.ncbs.res.in/DIAL/DIALserver.html) [21]. DIAL is an algorithm for domain identification in proteins where proteins are considered as a string of substructures (secondary structures and connecting loops) and the substructures are clustered according to their proximity indices. Each cluster, therefore, derived is a potential structural domain and disjoint factor is calculated for each domain organisation. This factor is a measure of compactness of the sub-structural clusters. Disjoint factor values more than 1.0 are considered to provide acceptable solutions of possible structural domain architecture for the protein of interest [21]. Based on the percentage of residue occurrence in helical, strand and loop regions, DIAL classifies the domains into four classes such as all alpha class, all beta class, alpha beta class and few secondary structure classes.
Structure and function relationship
The generated structure of the catalase protein was further analyzed using Sequence Annotated by Structure (SAS) server [22] and ProFunc server [16], for the structure and function relationship, and all identified domains, respectively.
Results and Discussion
3D structure prediction, evaluation and validation
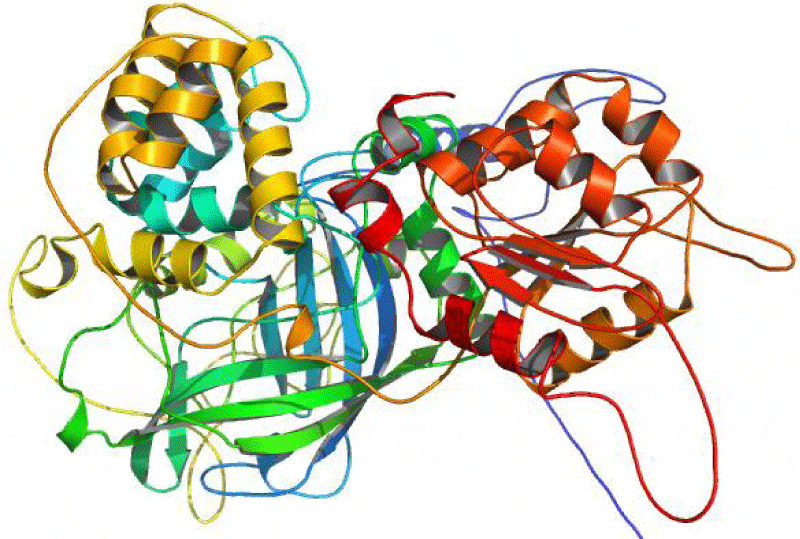
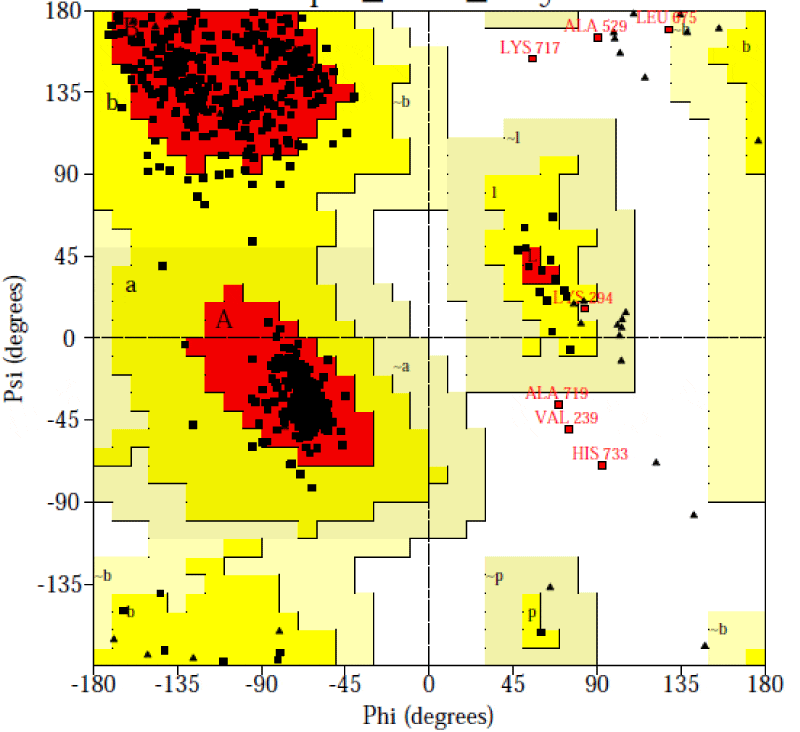
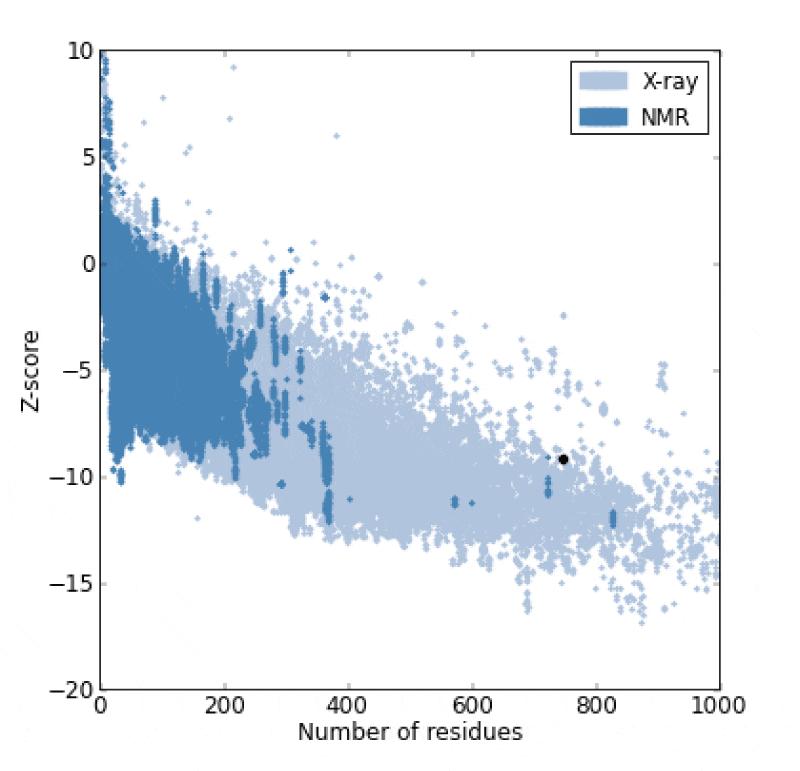
The catalase (CAT) is one of the most active enzymes (EC1.11.1.6) present in archea, eubacteria, fungi, plants, and animals. It dismutates hydrogen peroxide (H2O2) into one dioxygen (O2) and two water molecules [23]. Besides monophyletic monofunctional catalases, other enzymes, such as catalase-peroxidases, Mn-catalases, peroxidases and peroxiredoxins contribute to H2O2 disposal. H2O2 is inevitably formed in cells: most of it comes from superoxide (O2-) dismutation and from the activity of some oxidases [24,25]. Hence, a complete sequence of 749 amino acid residue for the A. fumigatus catalase enzyme was searched at the GenPept database and saved in fasta format (Accession no. AAB47761.1) It was reported that all the catalases share a highly conserved core structure but have nevertheless functional differences [26, 27]. Therefore, the homologous proteins with known structure were explored for the template selection of target protein in PDB database updated on 16 November 2013 using HHpred server. It was observed that the crystal structure of the catalase-1 from Neurospora crassa (PDB id: 1sy7_A) showed maximum similarity with the A. fumigates catalase selected as the template with the sequence similarity of 68% and E-value 5.1e-177. The predicted 3-D structure satisfied all the validation criteria on the basis of PROCHECK are illustrated in the Figure 1. According to the Ramachandran plot analysis of the predicted structure, 91.4% residues of Φ/Ψ angles are in the most favoured regions, 7.5% residues in the additional allowed region, 0.2% residues in generously allowed region and 0.9% residues in disallowed region as shown in the Figure 2. Additionally, the overall value of -0.08 was observed for G-factor, suggesting that the structure is highly unusual. Also, the ProSA server revealed that the model structure of the A. fumigatus catalase enzyme occupied the same region as observed in the X-ray predicted native protein structures with Z-score of −9.24 (Figure 3) It was also observed that the overall residue energies of the A. fumigatus catalase model were largely negative except for some peaks in few regions.
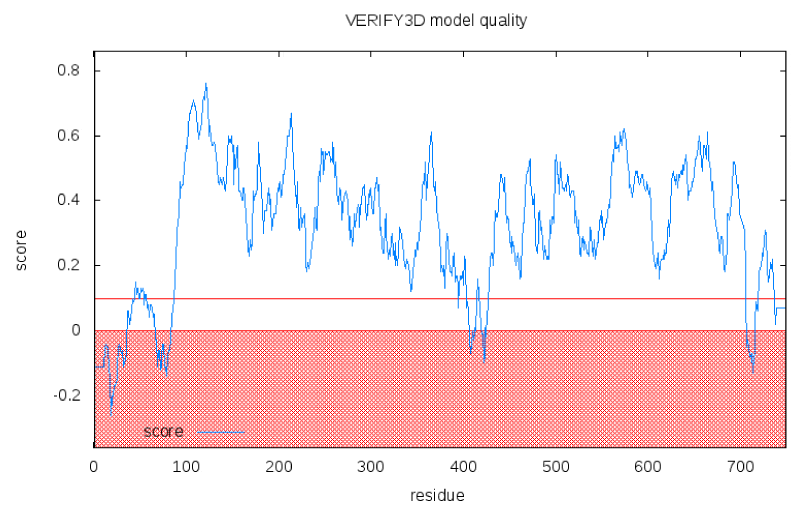
Besides, the results from the ProQ shows that the predicted structure is more reliable with LG score 3.77 and MaxSub 0.348. The overall quality for final structure has been further evaluated by Verify3D. The compatibility scores and the result for the final structure are presented in the Figure 4. The compatibility scores for all the residues in the developed model are above zero and hence, inferred that the generated 3D model for the catalase is reliable. The RMSD between predicted structure and template structure was found as 2.05A°, suggesting that the predicted 3D structure is an accurate model for the A. fumigatus catalase enzyme. The validated model was deposited in Protein Modeling Database with PMID PM0079726. Besides, with these evaluations, 3D model of A. fumigatus catalase was valid enough for high throughput virtual screening (VHTS) for designing potential antifungal drug.
Structural annotation and domain identification
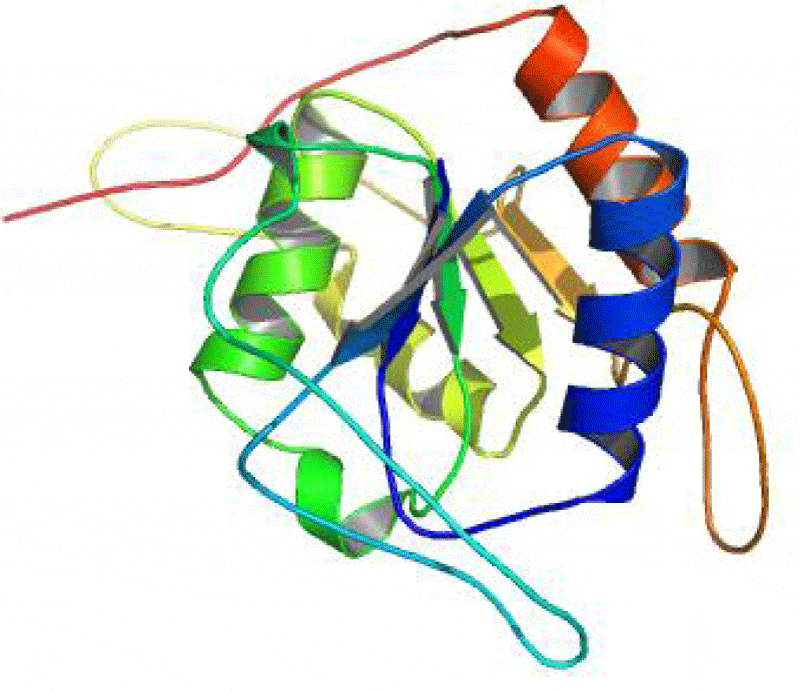
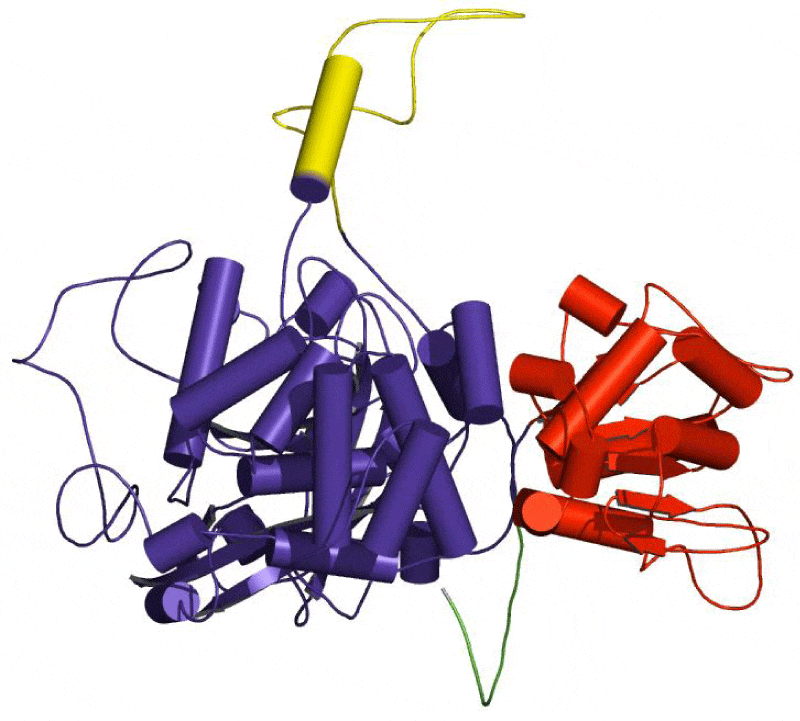
The SAS server was used to generate the wiring diagram as depicted in Figure 5. The ProMotif documentation of the enzyme via Profuncserver showed the results for the secondary structure summary as: the 749 residue span of the structure consists of 112 residues (15%), which were involved in the formation of the strands, 189 residues (25.2%) for the alpha helices, 26 residues (3.5%) for the 3–10 helix and 422 residues (56.3%) for the several other structural moieties. Also, the results show 4 beta sheets, 3 beta-alpha-beta motifs, 9 beta hairpins, 2 psi loops, 4 beta bulges, 21 strands, 30 helices, 29 helix-helix interactions, 64 beta turns and 9 gamma turns. Additionally, the four structural domains were also identified in the predicted 3D structure. The first domain related to few secondary structures class comprising 18 residues were detected from Met1 to Asn18, the second domain belonging to Alpha Beta class involves 497 residues was observed from Thr19 to His37 and Glu85 to Arg562, the third domain related to few secondary structures class consisting of 47 residues were established from Thr38 to His84 and the fourth domain belonging to Alpha Beta class consisting of 187 residues were found from Ala563 to Phe749. The 3-D structures of these domains are depicted in the Figures 6-9, respectively.
Structure and function relationship
Protein structure-function relationships can be investigated by asking how nature has reengineered protein structures to perform a variety of functions. Computational methods directed at the identification and analysis of related protein structures are an important prerequisite in this endeavour [28,29]. The structural domain regions were highlighted in predicted structure (Figure 10) SAS server reported three catalytic residues His93, Ser132 and Asn166 in the predicted structure while the positions of these catalytic residues are completely different in the template structure (His92, Ser131 and Asn165) [30]. This suggests that there is an insertion type mutation in the Aspergillus Fumigatus catalase when compared with Neurospora crassa catalase-1. The catalytic residues of the predicted structure were observed to be the part of second structural domain. The residues His 93 and Asn166 were found to form contact with hydrogen peroxide and many ligands, while Ser132 was found to contact with protoporphyrin IX containing Fe. Lys274 and Ala460, the part of second structural domain and Ser685, Glu693, Trp694 and Val697, the part of fourth structural domain were found to contact calcium ion. His84 found to contact chloride ion which is the part of third structural domain.
Conclusion
The present study projected the 3D structure of Aspergillus Fumigatus catalase. The structure to function relationship was also established by identifying the functional role of important residues in the four different structural domains of the modeled structure. This study purposed to discover new ligand with therapeutic potential to treat the virulence nature of this fungus for controlling the respiratory diseases.
- Singh BP, Banerjee B, Kurup VP (2003) Aspergillus antigens associated with allergic bronchopulmonary aspergillosis. Front Biosci 8: s102-109. Link: https://goo.gl/6bfeif
- Beauvais A, Latgé JP (2001) Membrane and cell wall targets in Aspergillus Fumigatus. Drug Resistance Updates 4: 38-49. Link: https://goo.gl/WKECRq
- Brakhage AA, Langfelder K (2002) Menacing mold: the molecular biology of Aspergillus Fumigatus. Annual Reviews in Microbiology 56: 433-455. Link: https://goo.gl/tQdMXL
- Varga J, Tóth B (2003) Genetic variability and reproductive mode of Aspergillus Fumigatus. Infection, Genetics and Evolution 3: 3-17. Link: https://goo.gl/yiYHeO
- Tomee JF, Kauffman HF (2000) Putative virulence factors of Aspergillus Fumigatus. Clin Exp Allergy 30: 476-484. Link: https://goo.gl/HnvMgN
- Denning DW (1998) Invasive aspergillosis. Clinical infectious diseases 781-803. Link: https://goo.gl/VcNAmG
- Latge JP, Debeaupuis JP, Sarfati J, Diaquin M, Paris S (1992) Cell wall antigens in Aspergillus Fumigatus. Arch Med Res 24: 269-274. Link: https://goo.gl/vmCuFK
- Schaffner A, Douglas H, Braude A (1982) Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus: observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. Journal of Clinical Investigation 69: 617-631. Link: https://goo.gl/UUmq2H
- Diamond RD, Clark RA (1982) Damage to Aspergillus Fumigatus and Rhizopus oryzae hyphae by oxidative and nonoxidative microbicidal products of human neutrophils in vitro. Infection and Immunity 38: 487-495. Link: https://goo.gl/31B40s
- Diamond RD, Krzesicki R, Epstein B, Jao W (1978) Damage to hyphal forms of fungi by human leukocytes in vitro. A possible host defense mechanism in aspergillosis and mucormycosis. The American journal of pathology 91: 313-328. Link: https://goo.gl/1quDhO
- Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, et al (2003) Catalases of Aspergillus Fumigatus. Infect Immun 71: 3551-3562. Link: https://goo.gl/RJQdr6
- Calera JA, Paris S, Monod M, Hamilton AJ, Debeaupuis JP, et al. (1997) Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect Immun 65: 4718-4724. Link: https://goo.gl/OItErZ
- Hamilton AJ, Holdom MD (1999) Antioxidant systems in the pathogenic fungi of man and their role in virulence. Med Mycol 37: 375-389. Link: https://goo.gl/DiTURE
- Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244-W248. Link: https://goo.gl/oAqRK3
- DeLano W (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA Link: https://goo.gl/3KZ7ZY
- Laskowski RA, Watson JD, Thornton JM (2005) ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33: W89-W93. Link: https://goo.gl/FaK9G8
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids res 35: W407-W410. Link: https://goo.gl/m4stNU
- Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12: 1073-1086. Link: https://goo.gl/rYy04Y
- Eswar N, Eramian D, Webb B, Shen MY, Sali A (2008) Protein structure modeling with MODELLER. Structural proteomics: high-throughput methods 145-159. Link: https://goo.gl/ptQbX5
- Singh DB, Gupta MK, Kesharwani RK, Misra K (2013) Comparative docking and ADMET study of some curcumin derivatives and herbal congeners targeting β-amyloid. Network Modeling Analysis in Health Informatics and Bioinformatics 2: 13-27. Link: https://goo.gl/DWn3Pm
- Sowdhamini R, Blundell TL (1995) An automatic method involving cluster analysis of secondary structures for the identification of domains in proteins. Protein Sci 4: 506-520. Link: https://goo.gl/GndZzQ
- Milburn D, Laskowski RA, Thornton JM (1998) Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng 11: 855-859. Link: https://goo.gl/LD7uAH
- Díaz A, Horjales E, Rudiño-Piñera E, Arreola R, Hansberg W (2004) Unusual Cys-Tyr covalent bond in a large catalase. J Mol Biol 342: 971-985. Link: https://goo.gl/4w1Lwt
- von Ossowski I, Hausner G, Loewen PC (1993) Molecular evolutionary analysis based on the amino acid sequence of catalase. J Mol Evol 37: 71-76. Link: https://goo.gl/rUIScG
- Klotz MG, Klassen GR, Loewen PC (1997) Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol Biol Evol 14: 951-958. Link: https://goo.gl/tqBlJ2
- Bravo J, Fita I, Gouet P, Jouve HM, Melik-Adamyan W, et al. (1997) Structure of catalases. Cold Spring Harbor Monograph Archive 34: 407-445. Link: https://goo.gl/DQkq3I
- Nicholls P, Fita I, Loewen PC (2000) Enzymology and structure of catalases. Advances in Inorganic Chemistry 51: 51-106. Link: https://goo.gl/4A4HYB
- Babbitt PC (1998) The relations hip between protein s tructure and function, or how have proteins over time diverged in function? Pacific Symposium on Biocomputing. PSB Proceedings 3: 459-460. Link:
- Kumar S, Bhagabati P, Sachan R, Kaushik AC, Dwivedi VD (2015) In Silico Analysis of Sequence–Structure–Function Relationship of the Escherichia coli Methionine Synthase. Interdiscip Sci 7: 382-390. Link: https://goo.gl/CpYkUh
- Díaz A, Horjales E, Rudiño-Piñera E, Arreola R, Hansberg W (2004) Unusual Cys-Tyr covalent bond in a large catalase. J Mol Biol 342: 971-985.. Link: https://goo.gl/tEQ1em
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
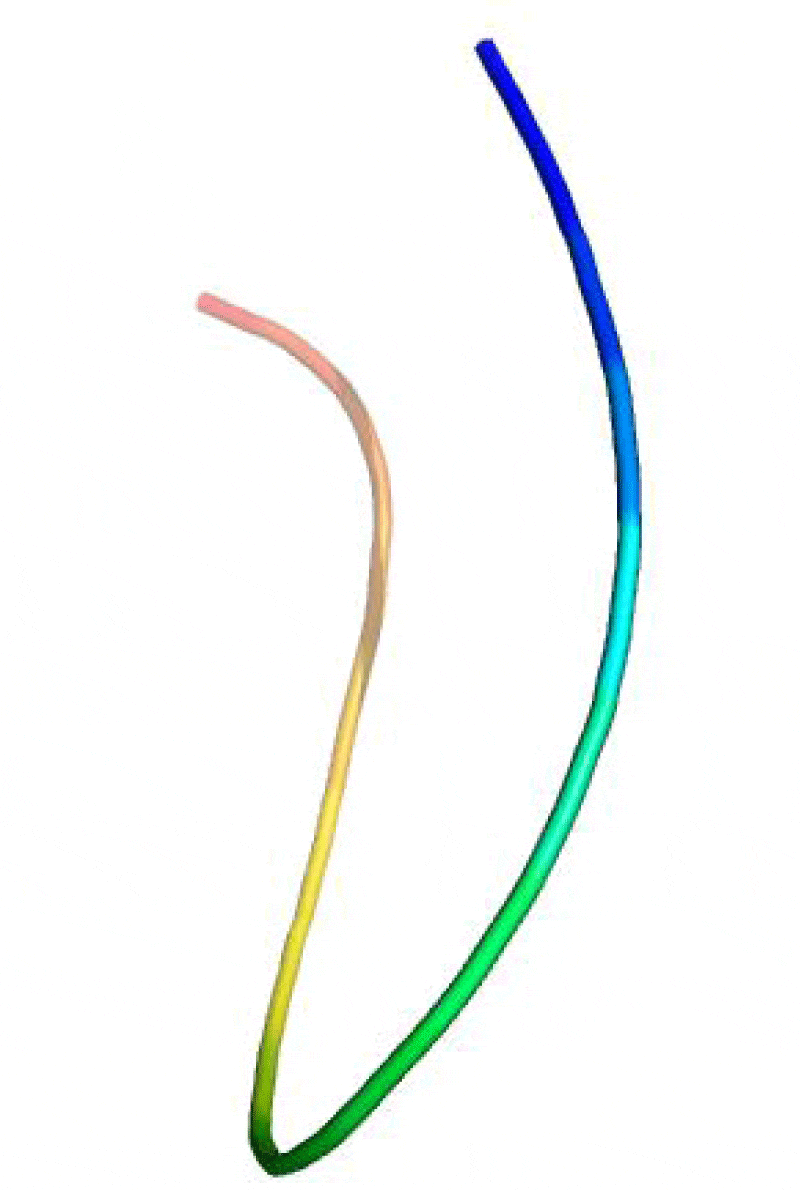

 Save to Mendeley
Save to Mendeley
