Open Journal of Bioinformatics and Biostatistics
A Preliminary analysis of potential allergens in a GMO Rice: A Bioinformatics approach
Custer C Deocaris1,2*, Rowena Grace Rumbaoa3, Anna Mae Gavarra4 and Malona V Alinsug5
2Technological Institute of the Philippines, Cubao Quezon City, Philippines
3Department of Food Science and Nutrition, College of Home Economics, University of the Philippines Diliman, Quezon City, Philippines
4Central Bicol State University of Agriculture, Pili, Camarines Sur, Philippines
5Science Department, College of Natural Sciences & Mathematics, Mindanao State University-General Santos City, Philippines
Cite this as
Deocaris CC, Rumbaoa RG, Gavarra AM, Alinsug MV (2020) A Preliminary analysis of potential allergens in a GMO Rice: A Bioinformatics approach. Open J Bioinform Biostat 4(1): 012-016. DOI: 10.17352/ojbb.000007This study uses an in silico approach in screening nascent allergens in GMO and conventionally-bred rice. The protein sequences analyzed were taken from published microarray data from GMO and conventionally-bred rice. To determine the proteins’ potential allergenicity, we used allergen databases and algorithms such as Allermatch and Algpred. Our analysis revealed the following putative allergenic proteins in GMO rice, namely: a cysteine proteinase precursor, a putative germin A, a glycosyl hydrolase, a subtilisin-like serine proteinase and an unknown protein. These genes are related to stress and defense response, metabolism, and moving, modifying, storing, and degradation of proteins.
Introduction
Large protein molecules, called allergens, frequently trigger an allergy. It involves a series of intrinsic and extrinsic reactions that contribute to triggering the symptoms (Somvanshi, et al., 2008). A typical allergy (Type I hypersensitive reaction) is induced by allergens that trigger specific IgE antibodies between common homologous allergens from different sources. Allergy symptoms include asthma, atopic dermatitis, and rhinitis, but severe reactions may also occur like an anaphylactic shock, which could lead to death.
Over half of the world’s population consumes rice as a staple food because it contains carbohydrates and proteins as energy sources. With the advent of novel foods, the number of patients with allergy to some foods has been increasing in recent years, and rice is one of these foods. Symptoms of rice allergy are asthma (Arai et al., 1998; Hoffman, 1975; Shibasaki ,et al., 1979), diarrhea and vomiting (Cavataio, et al., 1996), and atopic dermatitis with ocular complications (Uchio, et al., 1998).
Developing new cultivars by agricultural biotechnology has paved the way for crops that can adapt to poor environmental conditions and increased nutritional properties. However, these can change the properties of proteins found in food, thus increasing its allergenic potential. The Codex Alimentarius Commission states that safety assessments of genetically-modified (GM) foods need to include an investigation of tendencies to provoke allergy that might result from gene insertion (Haslberger, 2003). Comprehensive analysis, such as bioinformatics, may be useful tools in assessing allergenicity of newly expressed proteins in GMOs versus conventionally-bred crops. It covers various bioinformatics methods available, including allergen databases and algorithms for the search of sequence identity of newly expressed proteins with known allergens and assessing the relevance of alignments observed (Kaiserlian, et al., 2010).
Chemical comparison of a new protein with known allergens is a useful method for assessing a protein’s allergenic potential. Proteins are made up of long chains of amino acids, but their allergenicity is determined by only a few residues which serve as a binding site for antibodies. The ability of a protein to induce both humoral and cellular Th2 immune responses, leading to the release of allergen-specific IgE and Th2 cytokines, is a measure of its allergenicity [1]. Such property is usually assessed by in vitro and in vivo tests. No protein can be classified as allergenic without at least the evidence of the ability to bind in vitro IgEs from sera of sensitized individuals. If antibodies can attach themselves to a new protein and elicit a hypersensitivity cascade, there is a high chance that an allergic reaction can occur. The amino acid sequences of a protein are compared with the known allergens to determine homologies. A minimal requirement for sequence homology with a known allergen is a 35% sequence identity over a window of at least 80 amino acids [2].
GM crops undergo rigorous assessment for food, feed, and environmental safety before commercialization. However, detecting potential allergens, either in vivo or in vitro using molecular biology techniques, is challenging, time-consuming, and costly [3]. Additionally, eliciting an immune response is very complicated as the body responds to allergens by inducing many processes [4]. Possible development of potentially life-threatening allergic reactions or even the lack of sensitivity due to genetic factors could be some of these methods’ challenges. These limitations make the in silico (or bioinformatics) an acceptable, but a preliminary approach for identifying cross-reactive epitopes and allergenicity [5].
The allergenicity assessment for GM plants includes two approaches: (1) the assessment of the entire GM plant, and (2) the evaluation of the newly expressed proteins [6]. Based on the latter, in this study, an in silico based allergenicity screening is demonstrated as a quick and straightforward approach to identify potential allergens from newly expressed proteins based on differentially-expressed genes that arise from a transgenic variety of rice.
Materials and methods
Microarray data information
The protein sequences used in this study were obtained from the differentially expressed genes from the rice microarray data by Batista et al. [7]. Briefly, the gene expression profiles of GSE12069 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/). The dataset was based on the GPL2025 platform (Affymetrix Rice Genome Array, Thermo Fisher Scientific, Inc., Waltham, MA, USA). GSE12069 included data from a well-characterized transgenic rice line (cv. Bengal) and its non-transgenic mother plant as control (submission date, 10 July 2008). The stable transgenic line is on its third generation of self-pollination after transformation. The plant expresses a ScFV antibody (ScFvT84.66) against the carcinoembryonic antigen [7].
We used the GEO2R tool (http://www.ncbi.nlm.nih.gov/geo/geo2r/) to identify differentially expressed mRNAs from the GEO series [8]. Each sample within a GEO series was first classified into either normal or mutant variety. Then the defined groups were inputted into GEO2R. GEO2R provided a list of DEGs ranked according to differential expression levels. DEGs up-regulated significantly (>2.0-fold) and with significance (p < 0.01) were collected. Finally, the sequences were extracted from the David Functional Annotation Bioinformatics Microarray Analysis (http://david.abcc.ncifcrf.gov). The sequences analyzed in this study can be found in the DAVID Bioinformatics database with the following Accession Numbers: Q7F3A8, P0C5A4, Q6YZA4, Q0JDD4, Q0E256, Q7XVQ2, Q0IYV1, Q65XV2, Q0D652, Q6Z127, Q6Z127, Q6Z102, Q6K6Q0, Q75WV3, Q0INZ2, Q0JEB7, Q7XEZ6, Q0J525, Q6Z7P9, B7E541, Q6F2N5, Q6K1S6, Q53JL2, Q8H7P2, Q5N818, Q6YUS6, Q0J294, Q2R0M8, Q2R1H2, Q0IRH6, Q7XES5, Q2QXJ4, Q6H5C7, Q6PU50, Q0DVR9, Q10QUO, Q0DM56, Q7XUK3, Q0JBU0, Q0JA08, Q7XIE4, Q6Z4MO, Q8LIR5, Q6H421, Q0IYV1, Q33AW3, Q53PZ7, Q0ITQ0, Q0IQ30, Q2QNX3. The FASTA protein sequences of the differentially expressed genes were obtained from UniProt database (https://www.uniprot.org/) using Oryza sativa subsp. japonica [39947] as background.
In silico prediction of allergenicity
The protein sequences were subjected to allergenicity screening using AlgPred and Allermatch. AlgPred is an online tool that allows the prediction of allergens based on the similarity of known epitope with any region of a query protein (http://www.imtech.res.in/raghava/algpred/). Three modules were used: the SVM module based on amino acid composition prediction, the MEME/MAST motif prediction approach, and mapping of IgE epitopes. AlgPred allows the mapping of IgE epitope(s) on epitope in a given protein. It has a search function for MEME/MAST allergen motifs using MAST, which assigns if a protein is a possible allergen. MEME (Multiple Em for Motif Elicitation) is a tool for discovering motifs in related protein sequences. MAST (Motif Alignment and Search Tool) is a tool for searching biological sequence databases for sequences that contain one or more of a group of known motifs. AlgPred also allows the prediction of allergenicity based on SVM modules using amino acid or dipeptide composition. SVM is implemented using SVM_light [9] with input vectors on amino acid composition (20 vectors) and dipeptide composition (400 vectors) of each protein sequence. Finally, AlgPred facilitated a BLAST search against 2890 antigen-related proteins (ARPs) obtained and assign a protein allergen if it has a BLAST hit [10,11].
Allermatch is a FASTA package (version 3.4t21) available at ftp://ftp.virginia.edu/pub/fasta/. The default parameters (ktup = 2, matrix = Blosum50, Gap open = -10, Gap extend = -2). According to the current FAO/WHO Codex Alimentarius guidelines, the analysis tool provides the search methods where the alignment of 80-amino-acid sub-sequences of the input sequence was done using a sliding window of 80-amino-acid size (Fiers et al., 2004).
Homology modeling and visualization
Homology models of proteins are of great interest for better appreciating biological properties experiments when no experimental three-dimensional structures are available, especially if these are novel proteins. The 3D protein structure was modeled on the SWISS-MODEL workspace using the Alignment Mode [12], and was visualized using the program PyMol [13].
Results and discussion
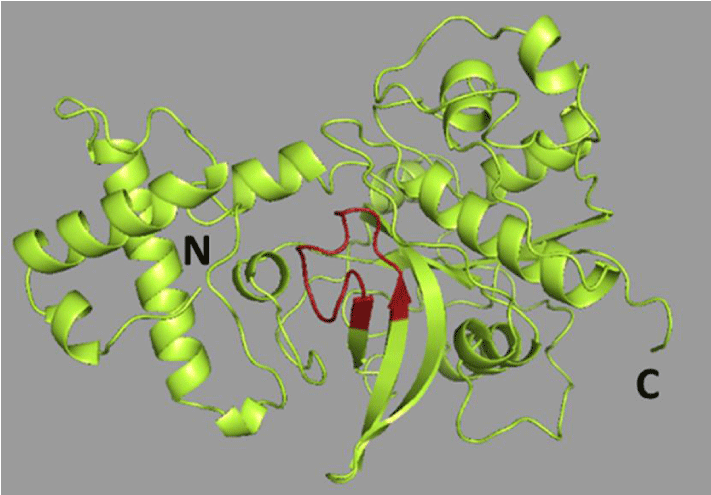
Following our preliminary screening of the 50 rice proteins with AlgPred and Allermatch, we found five proteins that were considered potentially allergenic (Table I). A cysteine proteinase precursor (Q7F3A8) was predicted to be the most allergenic. It contains the IgE epitope, VKNSWGTAWGEGGYI, which is homologous to known allergens from short ragweed (Ambrosia artemisiifolia), kiwi (Actinidia deliciosa), papaya (Carica papaya), pineapple (Ananas comosus), and soybean (Glycine max). A homology model of the protein is shown in Figure 1, and the IgE cross-reactive epitope is highlighted in red. Cysteine proteinase, one of the significant groups of plant proteases, has been thoroughly studied due to its crucial role in senescence and programmed cell death [14]. Interestingly, the potential allergen is a member of the structurally-related protein superfamilies - the prolamins [15]. These protein families are have structural domains of plant food allergens known to trigger a reaction via the gastrointestinal tract [16].
Based on the SVM method, due to dipeptide composition, a putative germin A protein (Q6YZA4) was predicted to be allergenic by AlgPred. It should be noted that the sensitivity, specificity, accuracy, and Mathew’s correlation coefficient for SVM for dipeptide composition for Algpred are 88.8%, 88.2%, 88.5%, and 0.770, respectively [10,17]. Germins and the related germin-like proteins (GLPs) are glycoproteins expressed in many plants in response to biotic and abiotic stress. In an immunoblotting assay, 24 out of 82 tested sera (29.26%) from allergic patients showed IgE-binding to germins. Germins can bind to IgE antibodies likely via their carbohydrate moieties [18].
Similar to germins, subtilisin-like serine proteinase (Q0E256) are also involved in the protective signaling mechanisms. ALE1, a gene homologous to subtilisin-like serine proteases, was found to be expressed within specific endosperm cells adjacent to the embryo and regulates the formation of cuticle on embryos and juvenile plants [19]. Strangely, its biological role in modifying proteins for plants is also mirrored by the C. elegans (nematode). A homolog subtilisin-like serine protein is involved in cuticle formation and essential for early development and adult morphology [20]. Subtilisin-like serine proteinase (Q0E256) in rice is homologous to the allergen Cuc m 1 from muskmelon. The thermally-stable Cuc m 1 is the only plant food allergen belonging to the family of serine proteases. Most allergens from this family are fungal allergens from the subfamilies of alkaline or vacuolar serine proteases [21].
The glucan endo-1,3-beta-glucosidase 14 (Q0JDD4), a poly-galacturonase, showed significant identity with allergen Hev b 2 (1,3-glucanase) of Hevea brasilienses (Para rubber tree) and Mus a 5 of Musa acuminate (banana). Hev b 2 is one of the allergens that cause latex allergy, an IgE-mediated hypersensitivity disorder in which patients are sensitized to natural rubber latex [22]. Recently, a novel allergen, PR-1a was cloned. PR-1a is a causative agent of peach tree pollen sensitization and is similar to glucan endo-1,3-beta-glucosidase 14. IgE of subjects who developed peach tree pollen allergies living in areas where this tree is widely cultivated was found to recognize a glucan endo-1,3-beta-glucosidase-like protein [23].
Lastly, we found potentially allergenic an unnamed protein (Q7XVQ2) that was overly expressed up to 19-fold in the transcriptome of the transgenic rice analyzed. A PFAM analysis indicated that it has an expansin C-terminal domain, a component of a plant cell wall protein involved in the non-enzymatic rearrangement of cell walls during cell growth. The expansin domain is associated with the allergens lol PI, PII, and PIII from Lolium perenne or ryegrass [24]. These grass pollens are widely implicated in the cross-sensitivity of people to house dust mites triggering severe asthma or allergic rhinitis [25].
From 1992-2017, more than 1,300 separate assessments by regulatory agencies around the world have reviewed the safety data on various GM crops, with every report concluding that the GM crop is as safe as conventionally-developed crops. For the 20 years of accumulated data, it has been concluded by far that the application of transgenic methods does not affect the levels of allergenic proteins native to a particular crop [6]. While a motif, sequence homology-based screening, is a part of a pipeline for allergenicity assessment of novel foods, we emphasize that in silico analysis should be backed up or validated by empirical research. A significant limitation of this study is that the potential allergens were data mined from transcriptome data. The food allergens Ara h 1 (peanut), Pru p 3 (peach ) and Gly m 5 (soybean ) are usually present at relatively high amounts from 1,000 to 10,000 ppm [6,26]. By contrast, the putative allergens proteins identified in this study were associated with only 8-15 fold increased in mRNA expression as compared to the non-GMO counterpart. Thus, it may be orders of magnitude lower than the sensitizing level of typical food allergens. There is also a possibility that post-translational modifications may happen downstream and that the potentially allergenic proteins may susceptible to gastric digestion. Of note, the ability of a protein’s epitopes to survive gastric digestion is an essential characteristic of food allergens, while in vitro pepsin resistance tests remain an integral part of the weight-of-evidence approach to assessing the allergenic potential of any novel protein [27]. Further research can focus on studying these potential allergens in the food matrix since the context of the food ingestion can modify the conformational epitopes. This technical issue assumes significance and needs to be given some thought concerning novel foods’ safety assessment.
Conclusions and perspectives
In conclusion, an in silico method may be a useful tool for the initial prediction of potentially allergenic proteins provided gene expression data is available. Five potential allergens have been identified for a transgenic rice variety. As recent studies recommend, modern bioinformatics tools could serve as a preliminary yet robust approach to identifying allergenic proteins in food [28-32]. Since this is a preliminary study, further tests should be done to conclude the likelihood of allergenicity of the proteins.
Authors’ contribution
RGR and AMG collected and analyzed the data and wrote the initial draft of the manuscript; CCD and MVA validated the initial bioinformatics results, performed homology modeling, contributed to writing, and finalized the paper.
- Goodman RE, Chapman MD, Slater JE (2020) The Allergen: Sources, Extracts, and Molecules for Diagnosis of Allergic Disease. The Journal of Allergy and Clinical Immunology: In Practice 8: 2506-2514. Link: https://bit.ly/2ZIHAJM
- Li KB, Issac P, Krishnan A (2004) Predicting allergenic proteins using wavelet transform. Bioinformatics 20: 2572-2578. Link: https://bit.ly/2Rw8fFc
- Kirsch S, Fourdrilis S, Dobson R, Scippo ML, Maghuin-Rogister G, et al. (2009) Quantitative methods for food allergens: a review. Analytical and bioanalytical chemistry 395: 57-67. Link: https://bit.ly/2E8E3Nk
- Parenti MD, Santoro A, Del Rio A, Franceschi C (2019) Literature review in support of adjuvanticity/immunogenicity assessment of proteins. EFSA Supporting Publications 16: 1551E. Link: https://bit.ly/2ZJKX3f
- Ahmed A, Minhas K, Namood-e-Sahar OA, Khan FS (2010) In silico identification of potential American cockroach (Periplaneta americana) allergens. Iran J Public Health 39: 109. Link: https://bit.ly/32Bu3FF
- Dunn SE, Vicini JL, Glenn KC, Fleischer DM, Greenhawt MJ (2017) The allergenicity of genetically modified foods from genetically engineered crops: a narrative and systematic review. Ann Allergy Asthma Immunol 119: 214-222. e213. Link: https://bit.ly/2ZK1KTM
- Batista R, Saibo N, Lourenço T, Oliveira MM (2008) Microarray analyses reveal that plant mutagenesis may induce more transcriptomic changes than transgene insertion. Proceedings of the National Academy of Sciences 105: 3640-3645. Link: https://bit.ly/3iFyhlc
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. (2012) NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 41: D991-D995. Link: https://bit.ly/33yEyJq
- Joachims T (1999) Making large-scale svm learning. Practical Advances in Kernel Methods-Support Vector Learning. Link: https://bit.ly/3mqaaJT
- Saha S, Raghava GP (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34. Link: https://bit.ly/35L26x5
- Verma AK, Misra A, Subash S, Das M, Dwivedi PD (2011) Computational allergenicity prediction of transgenic proteins expressed in genetically modified crops. Immunopharmacol Immunotoxicol 33: 410-422. Link: https://bit.ly/2FNeotT
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, et al. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46: W296-W303. Link: https://bit.ly/3mEuV4C
- Yuan S, Chan HS, Filipek S, Vogel H (2016) PyMOL and Inkscape bridge the data and the data visualization. Structure 24: 2041-2042. Link: https://bit.ly/3msFHe0
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. The Plant Cell 11: 431-443. Link: https://bit.ly/35ICK32
- Mills ENC, Madsen C, Shewry PR, Wichers HJ (2003) Food allergens of plant origin—their molecular and evolutionary relationships. Trends in Food Science & Technology 14: 145-156. Link: https://bit.ly/3iGmp2n
- Breiteneder H, Radauer C (2004) A classification of plant food allergens. J Allergy Clin Immunol 113: 821-830. Link: https://bit.ly/2RzaszJ
- Zhang L, Huang Y, Zou Z, He Y, Chen X, et al. (2012) SORTALLER: predicting allergens using substantially optimized algorithm on allergen family featured peptides. Bioinformatics 28: 2178-2179. Link: https://bit.ly/2ZGHHpe
- Jensen-Jarolim E, Schmid B, Bernier F, Berna A, Kinaciyan T, et al. (2002) Allergologic exploration of germins and germin-like proteins, a new class of plant allergens. Allergy 57: 805-810. Link: https://bit.ly/2FFjGYX
- Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, et al. (2001) A subtilisin-like serine protease is required for epidermal surface formation in <em>Arabidopsis</em> embryos and juvenile plants. Development 128: 4681-4689.
- Thacker C, Peters K, Srayko M, Rose AM (1995) The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genea Dev 9: 956-971. Link: https://bit.ly/2ZN22JH
- Kapingidza AB, Pye SE, Hyduke N, Dolamore C, Pote S, et al. (2019) Comparative structural and thermal stability studies of Cuc m 2.0101, Art v 4.0101 and other allergenic profilins. Mol Immunol 114: 19-29. Link: https://bit.ly/33FQxEO
- Crepy MN (2016) Rubber: new allergens and preventive measures. European Journal of Dermatology 26: 523-530. Link: https://bit.ly/2RyOOM1
- Blanca M, Victorio Puche L, Garrido-Arandia M, Martin-Pedraza L, Romero Sahagún A, et al. (2020) Pru p 9, a new allergen eliciting respiratory symptoms in subjects sensitized to peach tree pollen. PLoS One 15: e0230010. Link: https://bit.ly/3iDupkZ
- Ansari AA, Shenbagamurthi P, Marsh DG (1989) Complete primary structure of a Lolium perenne (perennial rye grass) pollen allergen, Lol p III: comparison with known Lol p I and II sequences. Biochemistry 28: 8665-8670. Link: https://bit.ly/33zF8qd
- Rengganis I, Pakasi LS (2018) Sensitization to food and pollen allergens and their implication for travelers with respiratory allergies. IOP Conference Series: Materials Science and Engineering 434: 012330. Link: https://bit.ly/2ZIrr72
- Larocca M, Martelli G, Grossi G, Padula MC, Riccio P, et al. (2013) Peel LTP (Pru p 3)–the major allergen of peach–is methylated. A proteomic study. Food Chemistry 141: 2765-2771. Link: https://bit.ly/3izexjj
- Wang R, Wang Y, Edrington TC, Liu Z, Lee TC, et al. (2020) Presence of small resistant peptides from new in vitro digestion assays detected by liquid chromatography tandem mass spectrometry: An implication of allergenicity prediction of novel proteins? PLoS One 15: e0233745. Link: https://bit.ly/2FD2TWf
- Blaauboer BJ, Boobis AR, Bradford B, Cockburn A, Constable A, et al. (2016) Considering new methodologies in strategies for safety assessment of foods and food ingredients. Food and Chemical Toxicology 91: 19-35. Link: https://bit.ly/3kr5oKk
- Herman RA, Song P (2020) Allergen false-detection using official bioinformatic algorithms. GM Crops & Food 11: 93-96. Link: https://bit.ly/33Cc8xZ
- Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, et al. (2002) Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int Arch Allergy Immunol 128: 280-291. Link: https://bit.ly/3iQARow
- Song P, Herman RA, Kumpatla S (2017) Bioinformatics Approaches to Identifying the Cross-Reactive Allergenic Risk of Novel Food Proteins. Food Allergy: Methods of Detection and Clinical Studies. Link: https://bit.ly/33Fksxb
- Hileman RE, Silvanovich A, Goodman R, Rice EA, Holleschak G, et al. (2002) Bioinformatic Methods for Allergenicity Assessment Using a Comprehensive Allergen Database. International Archives of Allergy and Immunology 128: 280-291. Link: https://bit.ly/3hysaOo
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
