Open Journal of Bioinformatics and Biostatistics
Partial genomic analysis of spike-glycoprotein among Sudanese camels infected with Middle East Respiratory Syndrome Coronavirus (MERS-CoV)
Ibrahim HS1*, Kafi SK1 and Musa HA2
2Research Laboratory, Faculty of Medical Laboratory Science, National Ribat University, Sudan
Cite this as
Ibrahim HS, Kafi SK, Musa HA (2020) Partial genomic analysis of spike-glycoprotein among Sudanese camels infected with Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Open J Bioinform Biostat 4(1): 006-011. DOI: 10.17352/ojbb.000006MERS-CoV was emerged for the first time in KSA; 2012 followed by a lot of new registered cases in the Middle East, European, American and African countries. The goal of this paper is to make a comparison between the partial spike-glycoprotein reference gene (NC_019843.3; genome region: 23864-24909 with 1046bp) with similar Sudanese camel sequences; using the NCBI data base. Creations of phylogenetic tree for those who were selected as reference genes from Middle East countries (Qatar, UKE, and KSA), south East Asia and African countries (Ethiopia, Egypt, Nigeria, Burkina Faso) with Sudanese sequences using ViPR website was done.
This study revealed that most of the Sudanese sequences descending from each other’s with a lot of mutations area which mainly concentrated at positions; 155, 156, 158, 242, 840, 876 at nucleotide levels and to explain these more in-depth studies are recommended.
Introduction
In the late 2012 the transmission of new virus that originated from bat – camel, called MERS-CoV was reported in Saudi Arabia when the first case of human infection was register followed by many reported cases in different countries [1-3].
Viral infections appear as mild cold flu; fever, cough, headache, myalgia, fatigue, shortness of breath and diarrhea and become severe with those with underline conditions [1,3,4].
According to genome phylogeny the virus classified to; clades A the first cases of viral infections (EMC/2012 and Jordan-N3/2012), clade B the second new cases of it; that shows genetically difference and clade C from Africa [1,5].
At that times many suggestions, studies and researchers appear on the top and leads to investigate African camels as source of the virus; one of this study was conducted in Egypt by Ali et al, 2017 that shown a higher seropositivity rate of MERS-CoV in camels imported from Sudan, Somalia and Ethiopia when camels sera were testing [6].
From 2016-2018 MERS-CoV viral screening among Sudanese camels was done and the partial genomic sequences of the spike glycoprotein were deposited in the gene bank [3].
The aim of this paper is to review the spike-glycoprotein sequences of Sudanese camels that retrieved from NCBI with that found in the others African and Asian countries to study the similarity and dissimilarity between them.
Materials and methods
The sequences of Sudanese camels spike-glycoprotein (1046pb length) were retrieved from NCBI website (which represented 89 sequences in addition to the other 22 reference sequences; with those gene bank access numbers; NC_019843.3, KT368872.1, KT368873.1, KT368880.1, KT368881.1, KT368860.1, KP966104.1, KP405225.1, MK967708.1, MG923475.1, MG923472.1, MG923466.1, MG923467.1, MG923469.1, MG923470.1, MG923471.1, MH734114.1, MH734115.1, KU242423.1, KU242424.1, KJ650098.1, MN507638.1 (https://www.ncbi.nlm.nih.gov/).
Sequence ID: NC_019843.3 (region: 23864- 24909; 1046bp, human isolate) was selected because it’s represented the first isolated sequences of MERS-CoV; that was deposited in the gene bank while the others ref-sequences (Camel isolates) were randomly selected according to study objectives.
Bioedit Sequence Alignment Editor Version: 7.0.9.1 was used to generate multiple sequence alignment (MSA) with SNP detections which was confirmed further more with Clustal W (https://www.genome.jp/tools-bin/clustalw).
MERS-CoV reference sequences from different Asian and African countries as I mentioned above were subjected to multiple sequence alignment with the creation of phylogenetic tree using ViPR (Virus Pathogen Resource) Release Date: Mar 22, 2020 (https://www.viprbrc.org/brc/home.spg?decorator=vipr).
Results
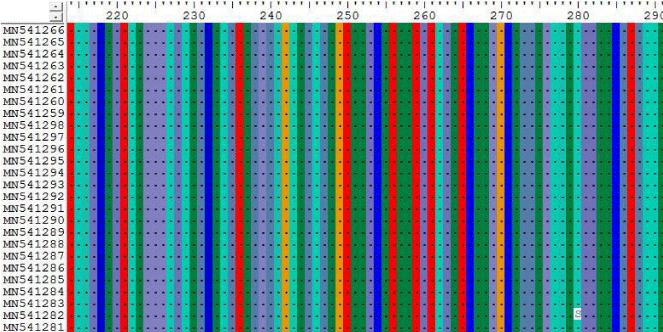
According to the MSA through Bioedit program and Clustal W, the following Sudanese camel sequences showed a variety of mutations when compared with ref-sequence (NC_019843.3) as illustrated below:
Nucleotide sequence
1- The results on Table-1 Shown the mutation at position 31and 54 for the following Sudanese camel sequences when compared with ref-sequence; NC_019843.3, see Figure 1).
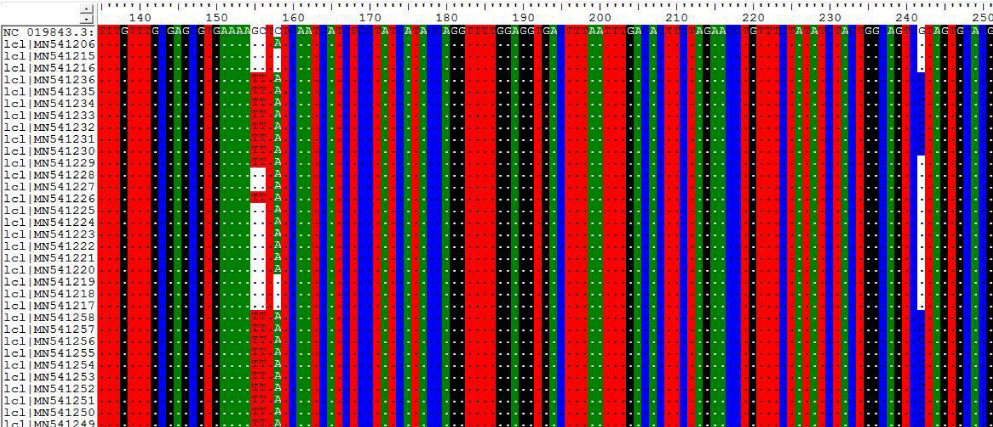
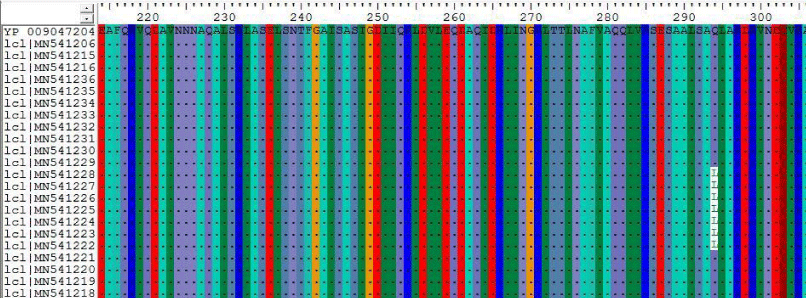
2- The results on Table-2 Shown the mutations at position 155,156,158 and 242 for the following Sudanese camel sequences when compared with ref-sequence; NC_019843.3, see Figure 2).
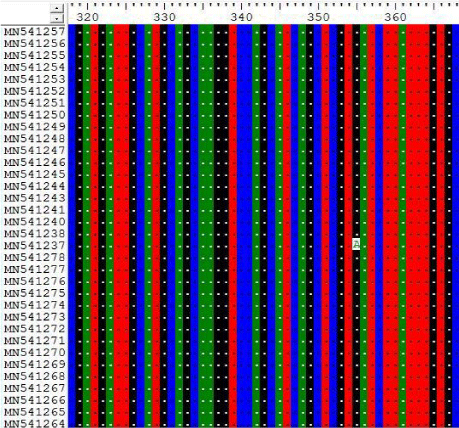
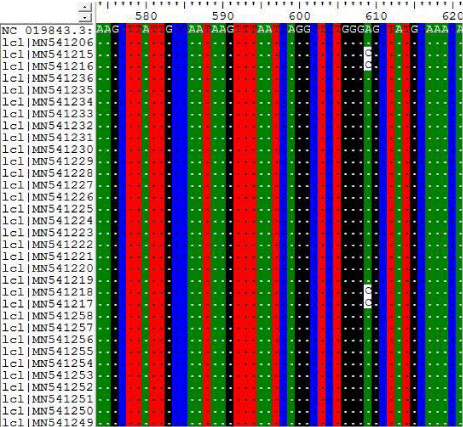
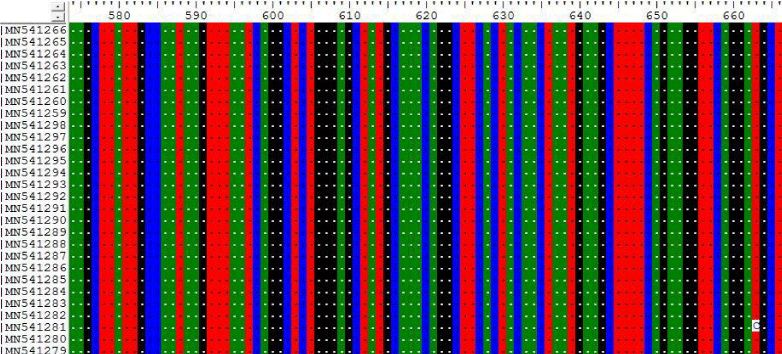
3- The results on Tables-3-6 Shown the mutations at position 355, 609, 663 and 838 on Sudanese camel sequences when compared with ref-sequence; NC_019843.3; see Figures 3- 6.
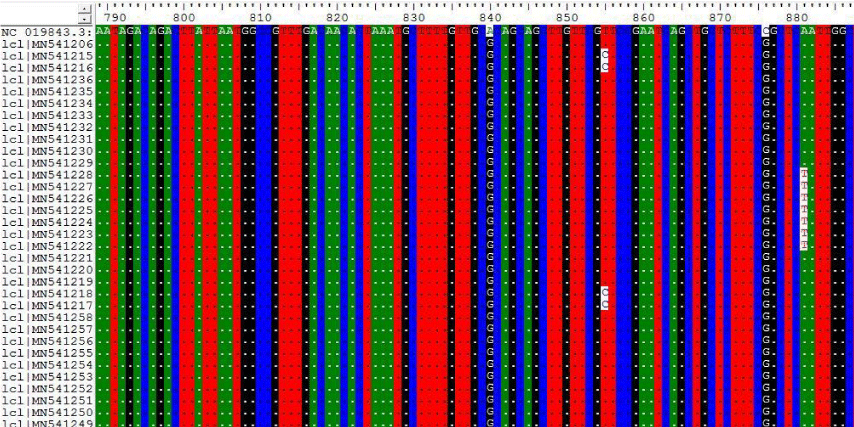
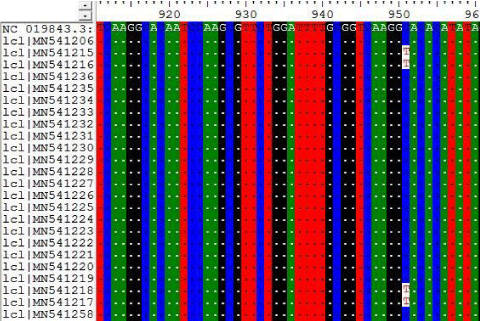
The results on Tables-7,8 shown the mutations at position 840, 855, 876, 801 and 951 for the following Sudanese camel sequences when compared with ref-sequence; NC_019843.3, see Figure 7,8).
Amino acid sequence
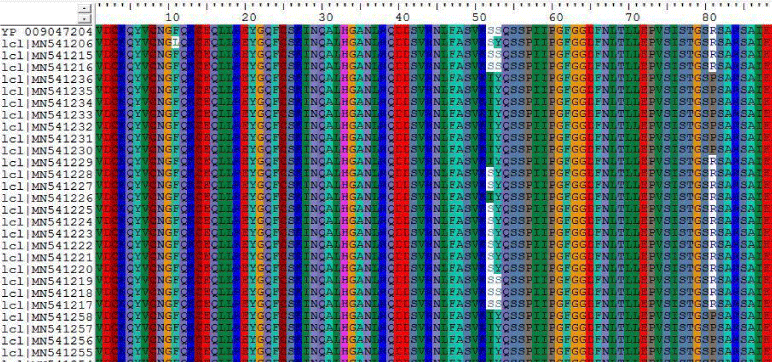
1- The results on Table 9 shown the conversion of amino acid at positions 11, 52, 53, 81 of Sudanese camel sequences when compared with ref-sequence; NC_019843.3, see Figure 9.
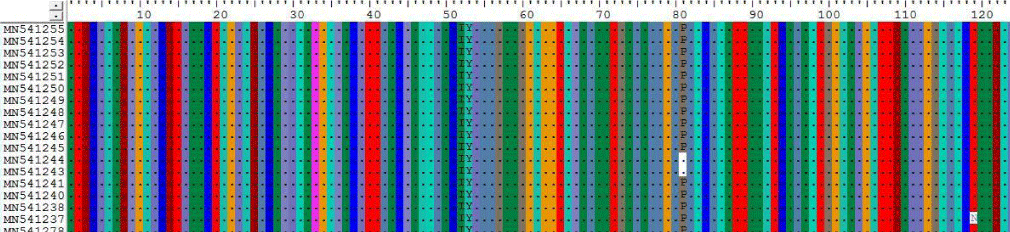
2- The results on table 10 shown the conversion of amino acid at positions 119, 294 and 280 for Sudanese camel sequences when compared with ref-sequence; NC_019843.3, see Figures 10-12.
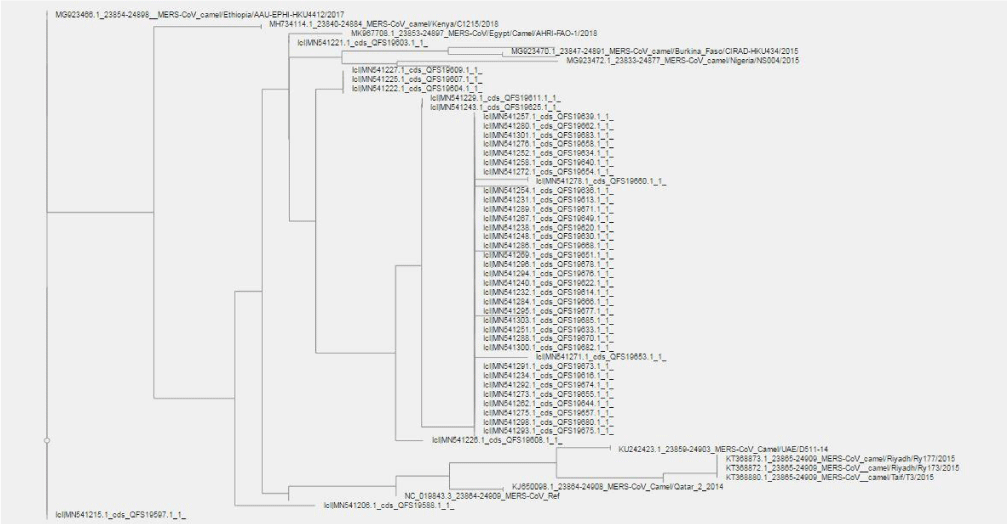
Phylogenetic analysis
89 MERS-CoV Sudanese sequences with 22 randomly selected ref-sequences with those gene bank accession numbers; NC_019843.3, KT368872.1, KT368873.1, KT368880.1, KT368881.1, KT368860.1, KP966104.1, KP405225.1, MK967708.1, MG923475.1, MG923472.1, MG923466.1, MG923467.1, MG923469.1, MG923470.1, MG923471.1, MH734114.1, MH734115.1, KU242423.1, KU242424.1, KJ650098.1, MN507638.1 were used for building phylogenetic tree.
Discussion
In this paper the MERS-CoV virus originated from each other and there are a lots of seems to be a conserved variant regions at positions 155, 156, 158, 242, 840 when compared with the others regions; the most common variant that differ from the ref-sequence (NC_019843.3) which was occurs at two positions A840G and C871G.
Phylogenetic tree of the 111 MERS-CoV sequences that included a 22 ref-sequences from African and Asian countries showed to be originate from; Ethiopia (MG923466.1; 2017) and Sudan (MN541215.1) followed by Kenya (MH734114.1; 2018) while Taif (KT368880.1; 2015) and Riyadh (KT368873.1, KT368872.1; 2015) sequences were similar to each other and had the same origin ancestor with the ref-sequence (NC_019843.3) that also contains; Qatar (KJ650098.1; 2014) and UAE (KU242423.1). Egypt strains (MK967708.1; 2018) seems to be closed to Sudan (MN541221.1); and this the last one divided into two branches one of them related to Nigeria (MG923472.1; 2015) and the other to Burkina Faso (MG923470.1; 2015); the remaining of Sudanese sequences shared some kind of similarity between each branches except Sudan (MN541226.1) sequence, see Figure 13).
Many studies indicated that MERS-CoV antibodies were early detected on camels mainly related to 1992 or earlier such as the study conducted between 2010 – 2013 on 310 dromedary camel that detected a higher neutralizing antibodies titer for the virus in addition RNA viral detection from nasal swabs (KSA) indicate sequence identity between it (some of them present some genomic variant with nasopharynx samples) and previously sequenced human isolates; the infections on young camels (≤ 2 years) considered more acute than adult and the risk of human infections increased in breeding seasons; the seropositivity is high while virus detection decreases [1,7,8].
In another study that studied the genetic and phenotypic characterization of MERS-CoV dromedaries samples in Morocco, Burkina Faso, Nigeria, and Ethiopia; illustrate the African virus was (clade C) which are phylogenetically distinct from the Arabian Peninsula (clades A and B) but it’s antigenically similar beside this the West Nigeria and Burkina Faso with North Morocco showed sub-clade C1 with the same deletions in the ORF4b gene; while C2 that contains multiple novel viruses in this study they are derived from Sudan and Djibouti as the viruses that previously detected by the authors and others in Egypt, Ethiopia, and Kenya; previously detected Egypt strain its designates a novel clade C3 [1,9,10].
Isolated MERS-CoV from Burkina Faso (BF785) and Nigeria (Nig1657) had lower virus replication in Calu-3 cells and in ex vivo cultures of human bronchus and lung; as BF785 shown lowered titer of replication in lungs of human DPP4-transduced mice. The reverse genetics-derived recombinant MERS-CoV (EMC) lacking ORF4b elicited higher type I and III IFN responses than the isogenic EMC virus in Calu-3 cells [9].
In El-Kafrawy S.A et al, 2019 study after taken 1196 nasal swabs from camels imported from Sudan (868) and Djibouti (328) into the Port of Jeddah in KSA for 2-year period (between Aug 10, 2016- May 3, 2018) and 472 samples from local Arabian camels over 2 months (May 1 and June 25, 2018) of which 189 were from Riyadh and 283 were from Jeddah, the study shows a higher prevalence of MERS-CoV in local camels than in imported camels (224 [47·5%] of 472 vs 157 [13·1%] of 1196; p<0·0001) which reached the peak on camels older than 1 year and aged up to 2 years in both groups, with 255 (66·9%) of 381 positive cases in this age group while the origin of West African strains in East Africa but East and West African strain was geographically separated and not re-sampled to Saudi Arabia despite sampling approximately 1 year after importation from Africa. All local Arabian samples related to virus strains that belong to a Novel Recombinant Clade (NRC) that first detected in 2014 in Saudi Arabia [10] Supplementary.
Conclusion
The results of this study showed that each new generation of Coronaviruses had mutations and different clade, we noticed that from different studies there is a deletion in ORF4b in MERS-CoV which showed recently in Africa; also Bat-SARS-CoV showed deletion on ORF8.
Transmissions of MERS-CoV from camel to human it’s still unclear and African data was limited beside the effect of divergent African culture.
This study indicated that Sudanese camel MERS-CoV virus sequences related to each other’s and had many mutations area. More in-depth studies are recommended.
The author would like to thank The National Ribat University member’s specially Prof. Shamsoun, Prof. Hassan and Dr. Mohammed.
- Middle East respiratory syndrome coronavirus From Wikipedia, the free encyclopedia (2020). Link: https://bit.ly/3jTNLmP
- Middle East respiratory syndrome coronavirus (MERS-CoV) – United Arab Emirates (2020). World Health Organization. Link: https://bit.ly/39GeK0j
- Middle East respiratory syndrome coronavirus (MERS-CoV) (2019). World Health Organization. Link: https://bit.ly/30dv5qd
- Killerby ME, Biggs HM, Midgley CM, Gerber SI, Watson JT (2020) Middle East Respiratory Syndrome Coronavirus Transmission. EID 26. Link: https://bit.ly/3fbceR6
- Chu Daniel KW, Hui Kenrie PY, Perera Ranawaka APM, Miguel E, Niemeyer D, et al. (2018) MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci U S A115: 3144-3149. Link: https://bit.ly/30aRFje
- Ali MA, Shehata MM, Gomaa MR, Kandeil A, El-Shesheny R, et al. (2017) Systematic, active surveillance for Middle East respiratory syndrome coronavirus in camels in Egypt. Emerg Microbes Infect 6: e1. Link: https://bit.ly/2P650De
- Reusken C, Haagmans BL, Koopmans MP (2014) Dromedary camels and Middle East respiratory syndrome: MERS coronavirus in the 'ship of the desert'. Ned Tijdschr Geneeskd. 158: A7806. Link: https://bit.ly/312ZWVS
- Dighe A, Jombart T, Van Kerkhove DM, Ferguson MN (2019) A systematic review of MERS-CoV seroprevalence and RNA prevalence in dromedary camels: Implications for animal vaccination. Epidemics 29: 100350. Link: https://bit.ly/3fcYd5n
- Daniel CKW, Kenrie HPY, Ranawaka APM, Eve M, Daniela N, et al. (2018) MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proceedings of the National Academy of Sciences of the United States of America 115: 3144-3149. Link: https://bit.ly/3hIN8L0
- El-Kafrawy SA, Corman VM, Tolah AM, Al Masaudi SB, Hassan AM, et al. (2019) Enzootic patterns of Middle East respiratory syndrome coronavirus in imported African and local Arabian dromedary camels: a prospective genomic study. The Lancet Planetary Health 3: e521-e528. Link: https://bit.ly/30d5VIz
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
