Open Journal of Bioinformatics and Biostatistics
Comparative modeling, structure and active site prediction of Sclerotinia disease-resistant gene BnaMPK3 in Oilseed rape (Brassica napus)
Zeshan Haider1, Muzdalfa Zulfiqar2, Iqrar Ahmad Rana1, Rao Sohail Ahmad Khan1, Muhammad Numan Akram3 and Adnan Khan Niazi1*
2Department of Bioinformatics and Biotechnology, GC University Faisalabad, Allama Iqbal Road Faisalabad, Punjab-38000, Pakistan
3Department of Neurology, Allied Hospital, Faisalabad Medical University, Faisalabad, Pakistan
Cite this as
Haider Z, Zulfiqar M, Rana IA, Ahmad Khan RS, Niazi AK, et al. (2020) Comparative modeling, structure and active site prediction of Sclerotinia disease-resistant gene BnaMPK3 in Oilseed rape (Brassica napus). Open J Bioinform Biostat 4(1): 001-005. DOI: 10.17352/ojbb.000005The prerequirement of genetic engineering is to have knowledge about structure and function of transgene and its encoded protein. This could be possible through in Insilco analysis of protein through protein comparative modeling which is a recent and important technique, used for the prediction of the protein three-dimensional (3D) structure. Fungal pathogen Sclerotinia sclerotiorum cause yield reduction about 15-20% in Brassica napus. Brassica napus mitogen-activated protein kinase 3 gene (BnaMPK3) codes for a BnaMPK3 protein that confers resistance against Sclerotinia sclerotiorum in oilseed rape. Computationnal analysis of primary structure, functional domains and S-Nitrosylation studies revealed that BnaMPK3 had five domains (protein kinases, kinases like superfamily, MAP kinase CS, Ser/Thr kinase AS, and protein ATP binding) and six S-Nitrosylation active sites at positions 53,141,176,181,227 and 336 respectively. The comparative modeling of the BnaMPK3 protein through Swiss-Model predicted 76.88% identity with template 6dtl.2A. 3-D structure of BnaMPK3 protein was validated through Errat score, which was 93.023%, indicated that it is stable protein. A total of 20 active sites were predicted by using ADDS (Automated active sites detection; docking; and scoring) software and active site with the highest overall quality score (ARG-72, ALA-77, ASP-115, VAL-151, ASP-155, LEU-156, ARG-173, PHE-174, GLY-175, ARG-178, PRO-179, THR-180, ARG-193, LEU-201, SER-205, and TYR-207) was used in further studies. The information achieved will be helpful to manipulate for sustainable resistance against sclerotinia disease in B. napus.
Introduction
Oilseed rape (Brassica napus L.) is an agronomically significant crop that is developed around the world; including; Europe; North America; and South Asia. The noteworthy requirement for its efficiency is sclerotinia disease brought about by fungal pathogen Sclerotinia sclerotiorum [1]. deBary; bringing about a huge misfortune in yield of seed: For instance; ten to twenty percent of yield misfortunes in a normal year; and round about eighty percent in seriously contaminated in fields of [2]. Surely, S. sclerotiorum is a massively damaging fungal plant pathogen which has host range of approximately four hundred eight plant species along with numerous economically significant crops. More than sixty names have been utilized to indicating the symptoms by this pathogen of fungal in horticulture. As of now; cross-breeding of S. sclerotiorum resistance cultivars of oilseed rape by utilizing conventional strategies is hard to manage; since no insusceptible or exceptionally resistance germplasm in Brassica napus is accessible [3]. The research activities are focused on activation of host defense system against S. sclerotiorum pathogen. Gene expression profile of host, in response to pathogen, demonstrated that genes related with jasmonic acid (JA) and ethylene(ET) signals are prompted. However, no Salicylic Acid (SA) related genes are found [4]. A few last investigations demonstrated that signaling of JA, together with SA and ET, adds to basal resistance against S. sclerotiorum [5]. These investigations summerise about the role of these defense effectors in protecting host from S. sclerotiorum, but not consistently. This information suggests that the activity of defense to the pathogen include various difficult organize of different signaling pathways.
It is reported that Mitogen-activated protein kinases (MAPK or MPK) assume significant function in the transduction of signals because of ecological stresses and hormones [6]. An action of MAPK constrained through consecutive actuation of two different kinases protein by which a MAPK kinase activates another MAPK kinase [7]. The proof established that some MAPK relatives have been involved as a part of protection signaling pathways in plant defense. For instance; in Arabidopsis thaliana; microbe-associated molecular patterns (e.g. MAMP, the bacterial flagellum-inferred flg22 peptide; stimulates the initiation of AtMAPK involving AtMPK1; AtMPK3; AtMPK4; AtMPK6; AtMPK11; and AtMPK13 [8]. AtMPK4 was first designated as a negative regulator of plant resistance [9]. Though some others, e.g. AtMPK3 was acknowledged to accept a positive role in plant defenseless dependent on their fast initiation through flg22 and pathogen immunization [10]. Furthermore, MPK3 mutant from the Arabidopsis thaliana work loss presented importantly less bacterial titters of Pseudomonas syringe; and the genes expression that controlled by SA synthesis and signaling; either handle or flg22-treated AtMPK3 loss of function mutants have no dierent from AtMPK4 mutants that appear upgraded protection from biotrophic fungal pathogens Hyaloperonospora arabidopsis’ and P. syringe [11]. In any case; another preliminary uncovered no reasonable vulnerable phenotype in the AtMPKs3 mutants when showering or infiltration-vaccinated with P. syringae [12]. MPK3 genes have been reported in different harvested species of plants and involved in defense responsive and additional biological procedures. In tobacco plant, WIPK (wound-induced protein kinase), the MPK3 ortholog in cultivated tobacco; was seen as activating during infection with (TMV) tobacco mosaic virus [13]. B. napus started from an unconstrained hybridization of B. rapa L. (2n = 20]) and B. oleracea L. (2n = 18). The yield is more complicated than that of the model plants Arabidopsis thaliana and Oryza sativa . Analysis of gene association based on various SNPs (Single Nucleotide Polymorphisms), which fluctuated over the controlled population, is used to dismember the locus commitments to the objective attributes and more to increase gene biological function. Infections brought about by S. sclerotium pathogen have customarily been very hard to manage the function [14]. In this investigation, we used the bioinformatics approach which is the best way to understand the function and characterize the BnaMPK3 protein. This article intended to 3D-model of the BnaMPK3 protein and anticipate its active binding site in-silico. The data accomplished will be utilized to distinguish the suitable effectors for BnaMPK3 protein first in silico and afterward in vitro.
Methodology
Amino acid sequence retrieval of BnaMPKs3
The sequence of the BnaMPK3 protein was obtained from the NCBI (National Center for Biotechnology Information); a large online (https://www.ncbi.nlm.nih.gov/) nucleotide sequence database (Accession # JQ708040.1).
Analysis of the primary structure and Factional domain of BnaMPK3
For the calculation of different chemical and physical characteristics of the BnaMPK3 protein 3D model, the ProtParam tool (http:// web.expasy.org/protparam/) was used. The online tool facilitated to determine the molecular weight, theoretical isoelectric point (pI), estimated half-life and instability index of the three-dimensional structure of BnaMPK3. The functional domain of BnaMPK3 protein sequences was determined by using the Interpro online tool available on the EBI web page (www.ebi.ac.uk/interpro/).
Computational analysis of S-Nitrosylation active sites in BnaMPK3 Protein
For the computational prediction of different S-Nitrosylation active sites in BnaMPK3 protein, the GPS-SNO ( http://sno.biocuckoo.org/.) bioinformatics tool was used. The prediction analysis of S-Nitrosylation active site through using GPS-SNO was well with good accuracy, specificity, and sensitivity, respectively. GPS-SNO predicted the reputed S-Nitrosylation sites of the different possibly S-Nitrosylated substrates for which the S-Nitrosylation sites were not predicted [15].
Comparative based modeling of BnaMPK3
Comparative modeling of BnaMPK3 proteins was done by utilizing the SWISS-MODEL web Bioinformatics tool (https://swissmodel.expasy.org/). It is an online web computational tool used for the homology modeling of protein three-dimensional structure [16].
Evaluation of the three-dimensional structure of BnaMPK3
The 3D computational structure of the BnaMPK3 protein was evaluated by ERRAT. This model examines the measurements of non-bonded connections among various sorts of atoms. This model additionally helpful to plot the estimation of the position of a 9-buildup sliding window versus the function’s ERROR. Out-puts were attained by comparing the values with statistically obtained after the refined structures [17].
Active site prediction of BnaMPK3
ADDS (Automated active sites detection; docking; and scoring) an online bioinformatics tool was used to predict active sites [18] of BnaMPK3 protein. The accuracy of this software was checked by the molecules of known active sites.
Result and discussion
Amino acid sequence retrieval of BnaMPK3
An amino acid sequence of BnaMPK3 protein (Accession # JQ708040.1) retrieved from NCBI was consisted of 370 amino acids.
Analysis of the primary structure and Factional domain of BnaMPK3
Analysis of the primary structure of BnaMPK3 was completed using ProtParam. BnaMPK3 has a total of 370 amino acids, molecular weight 42601.82 and theoretical pI 5.79. the instability index (II) was 36.39, which classified as stable protein. The estimated half-life for mammalian reticulocytes (in Vetro) was thirty hours, while in yeast and E. coli (In vivo) was greater than twenty and ten hours, respectively. The functional domains of BnMPK3 protein were predicted through the Interpro online server, which was further used for the construction of the three-dimensional structure of BnMPK3.
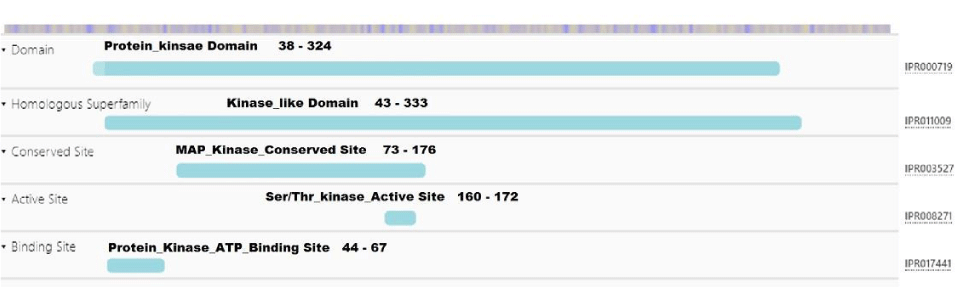
The Protein kinase domain (range 38-324 ), kinase-like domain superfamily (range 43 – 333), MAP Kinase conserved site ( range 73 – 176), Ser/Thr Kinase active site (range 160 – 172) and Protein Kinase ATP binding site (range 44–67), respectively; the five different functional domains of BnaMPK3 protein was predicted (Figure 1).
Computational analysis of S-Nitrosylation active sites in BnaMPK3 Protein
S-Nitrosylation plays an important character in biological functions. Therefore, the prediction of S-Nitrosylation active sites through using bioinformatics tools is efficient and quick. The estimated analysis showed six different S-Nitrosylation active site through using GPS-SNO was well with good accuracy, specificity and sensitivity, 75.80%, 53.57%, and 80.14%, respectively (Table 1).
Comparative based modeling of BnaMPK3 protein
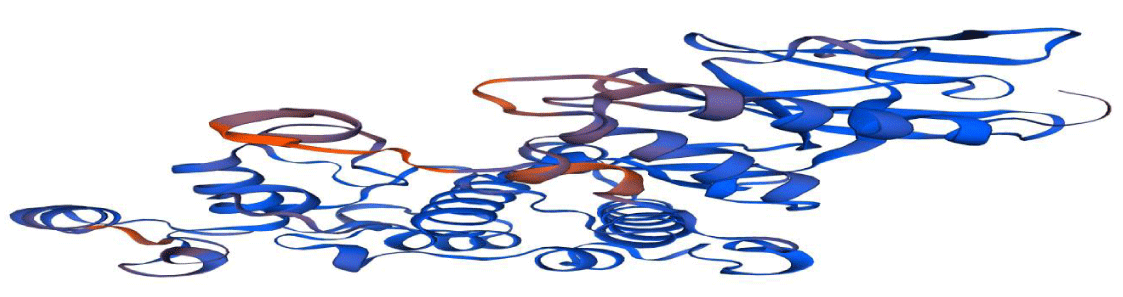
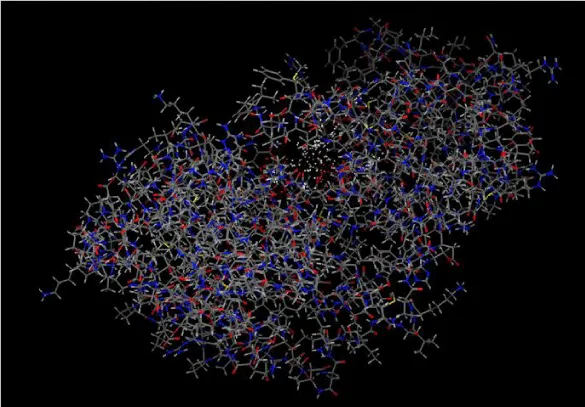
The comparative modeling of the BnaMPK3 protein of oilseed rape (Brassica napus); which is encoded by Sclerotinia disease-resistant gene BnaMPK3 was done by using a bioinformatics tool “SWISS-MODEL”. The 3D model was constructed using the amino acid sequence as a template for the BnaMPK3 protein. One best model was generated with maximum GMQE (Global Model Quality Estimation) from the Swiss model (Figure 2, Table 2).
Evaluation of the three-dimensional structure of BnaMPK3
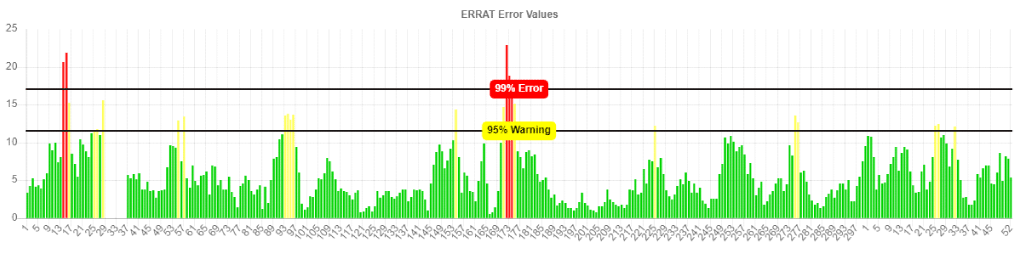
The obtained best 3D-model was evaluated by ERRAT. The best 3D-model of BnaMPK3 protein with a highly refined overall quality score 93.023%; which indicated the stability of the 3D-structure of BnMPK3 protein (Figure 3).
Active site prediction of BnaMPK3
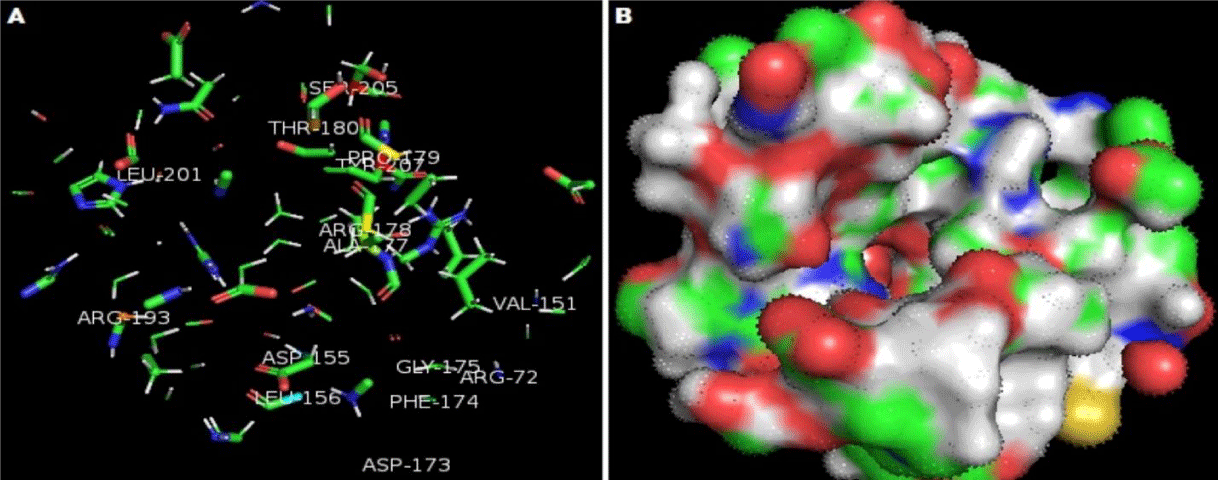
Active sites of oilseed rape (Brassica napus) BnaMPK3 protein was attained using ADDS (Automated active sites detection; docking; and scoring). A total of 20 active sites were predicted firstly (Figure 4). The only best pocket with the highest score was selected for further analysis (Figure 5). There are five functionals domains are identified in BnaMPK3 protein (38 – 324, 43 – 333, 73 – 176, 160 – 172, and 44 – 67). Protein kinases area engaged with cell flagging, multiplication, and advancement of the plant. The domain of protein kinases secured ABA-interceded reaction to injuring, osmotic pressure and pathogen assaults [19]. In plants MAPK tangled in the different developmental processes like cell signaling, differentiation, developments, and various kind of stress (abiotic and biotic) [20]. Mitogen-actuated kinases (MPKs) made an interpretation of the outer sign into the cell reaction that controlled cell passing in all eukaryotes. In plants, likewise two MPKs MPK3 and MPK6 are intricated to an alternate sort of organic procedures, for example, inborn immunity against the pathogen, cell motioning of various cell capacities, and development of plant however the component of these flagging are as yet not comprehended [21]. MPK6 was used as a template (76.88% identity) protein for prediction of the three-dimensional structure of BnaMPK3 Protein. The BnaMPK3 protein has five functional domains with the Leucine-Rich Region (LRR) and six S-Nitrosylation active sites make it effective against Sclerotinia sclerotiorum, LRR activated the innate immunity against pathogen so that known as fungus protein [22,23].
The active site with the highest score (ARG-72, ALA-77, ASP-115, VAL-151, ASP-155, LEU-156, ARG-173, PHE-174, GLY-175, ARG-178, PRO-179, THR-180, ARG-193, LEU-201, SER-205, and TYR-207) had the amino acids residues ARG-178, PRO-179, THR-180, ARG-193, LEU-201, SER-205, and TYR-207 were found within range of protein kinases and kinases domain of superfamily. While ARG-173, PHE-174, GLY-175 were found under the range of MAP kinases conserved domain and ARG-72, ALA-77, ASP-115, VAL-151, ASP-155, LEU-156, were found under the range of Ser/ Thr kinases active site domain. This indicated that BnaMPK3 protein has a high potential resistant gene Sclerotinia disease.
Conclusion
The main objective of this study is that to understand the Insilco biological function of BnaMPk3 protein and determined the active sites involved in resistance against Sclerotinia sclerotium pathogen. This bioinformatics analysis shows that BnaMPk3 protein is 370 amino acid long, that contains the major five different functional domains, protein kinases with range of 24 – 324, kinases like domain superfamily with range of 43 – 333, MAP kinases conserved domain with range of 73 – 176, Ser/Thr kinases active site domain with range of 160 – 172 and protein kinases ATP binding site domain with range of 44 – 67. These domains complicated in different biological functions in plants such as abiotic and biotic stress, differentiation and development of the plant. The predicted the computational based 3D structure of the BnaMPK3 by homology modeling showed 76.88% identity with template MPK6 (6dtl.2) and visualized with the assistance of online computational software. The active site of BnaMPK3 protein contained amino acid residues ARG-72, ALA-77, ASP-115, VAL-151, ASP-155, LEU-156, ARG-173, PHE-174, GLY-175, ARG-178, PRO-179, THR-180, ARG-193, LEU-201, SER-205, and TYR-207 respectively, the study of functional domains and computational analysis of BnaMPK3 will demonstrate to be a valuable model for exploiting in the future to understand the mechanism of resistance in BnaMPK3 against Sclerotinia disease.
Authors’ contributions
Conceived and designed the experiments: Iqrar Ahmad Rana, and Adnan Khan Niazi, Performed the experiments: Zeshan Haider; Analyzed the data: Adnan Khan Niazi and Rao Sohail Ahmad Khan; contributed materials/ analysis/ tools: Muzdalfa Zulfiqar and; Wrote the paper: Zeshan Haider and Adnan Khan Niazi.
- Lu G (2003) Engineering Sclerotinia sclerotiorum resistance in oilseed crops. Afr J Biotechnol 2: 509-516. Link: https://bit.ly/3dbtSmD
- Wu J, Cai C, Tu J, Li L, Liu S, et al. (2013) Identification of QTLs for resistance to Sclerotinia stem rot and BnaC. IGMT5. a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PloS One 8: e67740. Link: https://bit.ly/2CkOHiT
- Bolton MD, Thomma BP, Nelson BD (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7: 1-16. Link: https://bit.ly/3dc5DEZ
- Zhao J, Buchwaldt L, Roger Rimmer S, Sharpe A, McGregor L, et al. (2009) Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Molecular Plant Pathology 10: 635-649. Link: https://bit.ly/2YdnrLJ
- Nováková M, Sašek V, Dobrev PI, Valentová O, Burketová L (2014) Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum–Reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol Biochem 80: 308-317. Link: https://bit.ly/3deGTvA
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245-266. Link: https://bit.ly/2UWG5W6
- Rodriguez MCS, Petersen M, Mundy J (2010) Mitogen-Activated Protein Kinase Signaling in Plants. Annu Rev Plant Biol 61: 621-649. Link: https://bit.ly/2AO43Mb
- Nitta Y, Ding P, Zhang Y (2014) Identification of additional MAP kinases activated upon PAMP treatment. Plant Signal Behav 9: e976155. Link: https://bit.ly/2N7MvgJ
- Brodersen P, Petersen M, Nielse HB, Zhu S, Newman MA, et al. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J 47: 532-546. Link: https://bit.ly/2UTj6uQ
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci . 6: 520-527. Link: https://bit.ly/37E6P2R
- dit Frey NF, Garcia AV, Bigeard J, Zaag R, Bueso E, et al. (2014) Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol 15: R87. Link: https://bit.ly/2BfNCZc
- Su J, Zhang M, Zhang L, Sun T, Liu Y, et al. (2017) Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29: 526-542. Link: https://bit.ly/2YGiVnW
- Zhang S, Klessig DF (1998) The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc Natl Acad Sci U S A 95: 7225-7230. Link: https://bit.ly/2N8mpdp
- Takahashi Y, Teshima K, Yokoi S, Innan H, Shimamoto K (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences 106: 4555-4560. Link: https://bit.ly/3ei8Gg8
- Xue Y, Liu Z, Gao X, Jin C, Wen L, et al. (2010) GPS-SNO: Computational Prediction of Protein S-Nitrosylation Sites with a Modified GPS Algorithm. PLoS One 5: e11290. Link: https://bit.ly/2BhiisT
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195-201. Link: https://bit.ly/2Ck0Btj
- Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein sci 2: 1511-1519. Link: https://bit.ly/2ACvdWx
- Singh T, Biswas D, Jayaram B (2011) AADS-An automated active site identification, docking, and scoring protocol for protein targets based on physicochemical descriptors. J Chem Inf Model 51: 2515-2527. Link: https://bit.ly/3eeUJiS
- Laurie S, Halford NG (2001) The role of protein kinases in the regulation of plant growth and development. Plant Growth Regulation 34: 253-265. Link: https://bit.ly/3ddEvpd
- Ligterink W, Hirt H (2001) Mitogen-activated protein (MAP) kinase pathways in plants: Versatile signaling tools. in International Review of Cytology. Int Rev Cytol 201: 209-275. Link: https://bit.ly/2YcdGgD
- Putarjunan A, Ruble J, Srivastava A, Zhao C, Rychel AL, et al. (2019) Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to Speechless. Nature Plants 5: 742-754. Link: https://go.nature.com/3d7Pq3y
- Sun J, Steenbergen C, Murphy E (2006) S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal 8: 1693-1705. Link: https://bit.ly/3hz4ElH
- Shanmugam V (2005) Role of extracytoplasmic leucine rich repeat proteins in plant defence mechanisms. Microbiol Res 160: 83-94. Link: https://bit.ly/30ReWaQ
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
