Journal of Clinical Microbiology and Biochemical Technology
Isolating Lysobacter enzymogenes strains with enhanced protease activity via chemical mutagenesis
Malihe Amirkhani1,2, Vahideh Valizadeh2*, Maryam Davoudi2, Malihe Keramati2, Reza Ahangari Cohan2, Seyed Mohammad Atyabi2 and Dariush Norouzian2
2Nanobiotechnology Department, New Technologies Research Group, Pasteur Institute of Iran, Tehran, Iran
Cite this as
Amirkhani M, Valizadeh V, Davoudi M, Keramati M, Cohan RA, et al. (2022) Isolating Lysobacter enzymogenes strains with enhanced protease activity via chemical mutagenesis. J Clin Microbiol Biochem Technol 8(1): 010-015. DOI: 10.17352/jcmbt.000049Copyright
© 2022 Amirkhani M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Proteases are the most important industrial enzymes which have attracted enormous attention due to their vast variety and well-defined specificity. Microbial proteases are superior to other sources like plant and animal proteases because of their desired characteristics for biotechnological application. In this regard, Lysobacter enzymogenes is a rich source for the production of antibiotics and proteases. However, strain improvement in order to obtain overproduced microorganisms is always demanded at an industrial scale. Therefore, in the present study in order to enhance L. enzymogenes protease production, random mutagenesis was applied using N-methyl-N’-nitro-N-nitrosoguanidine (NTG) as a chemical mutagen. Random mutagenesis was conducted on L. enzymogenes suspension cultivated on nutrient broth using different concentrations of NTG (100, 150, and 200 µg/ml) for 20 and 40 minutes. The treated bacteria were cultivated on nutrient agar containing casein as a selective media. Primary and secondary screenings were performed by measuring the diameter of the casein hydrolysis zones in the isolated bacteria and the related supernatants, respectively. Finally, the unit of protease activity was quantified by Anson’s method of examining bacterial supernatants. Among the total of 30 isolated mutants, two mutants showed the highest level of extracellular proteolytic activity which showed 2.65 and 1.86 fold increments in contrast to the wild type, respectively. In general, the effect of mutagenesis by NTG can be emphasized to increase protease activity.
Introduction
Proteases, the largest group of hydrolytic enzymes account for approximately 60% of total worldwide marketed enzymes [1]. Since these enzymes play an important role in physiological processes, they can be extracted from immense sources such as animals, plants, and microorganisms [2]. However, microbial proteases are preferred for industrial applications due to the source availability and affordability, rapid growth rate, and ease of genetic manipulations [3,4]. Lysobacter enzymogenes which is known as a bio-control agent commonly produces many vital types of hydrolytic enzymes to control its environmental pathogens [5-8]. However, these bacteria release the enzyme only to the extent of their own need. Hence, for commercial purposes, strain improvement is mandatory to obtain mutant bacteria with increased enzyme production.
Among all natural proteases obtained from L. enzymogenes, Lysyl endopeptidase which specifically cuts proteins from the Lys-I-Xaa bond attracts more attention in biotechnology [9,10]. In addition to the lysine specificity, the most important feature of the enzyme is maintaining activity in the presence of denaturing agents such as urea and sodium dodecyl sulfate (SDS). These characteristics made the enzyme very useful in proteomics techniques including sequencing, analyzing, and determining protein structures (peptide mapping) and also in-gel digestion protocols [11].
Mutagenesis is the most commonly used approach for strain improvement that can be induced randomly or site-specific [12,13]. Random mutagenesis is a process in which, by using chemical and physical mutagenic agents, changes are made in the characteristics of products produced by genes. Sometimes the product is not produced at all or abnormal products are produced. But sometimes this process leads to increased harvest production compared to the non-mutant or parent strain [14,15]. In this regard, N-methyl-N’-nitro-N-nitrosoguanidine (NTG) as a classical DNA alkylating agent, is one of the most effective chemical mutagens that converts GC to AT in DNA strands, so it can generate a large number of mutants under optimum conditions [16]. Therefore, in the present study, random mutagenesis using NTG as a chemical mutagen was employed to enhance protease production of the L. enzymogenes strain. The isolated mutants with hyper proteolytic activity will be further analyzed in order to obtain industrial strain for the production of Lysyl endopeptidase.
Material and method
Microorganism and growth media
The bacterial strain Lysobacter enzymogenes 29486™ was purchased from American Type Culture Collection (ATCC, USA). The bacterium was revived in brain heart infusion broth (Merck, Germany) and incubated at 33 ± 1 °C based on ATCC recommendation. The medium was enriched for growth rate and protease production by adding 1% glucose, 0.01% of each mono-and dibasic potassium phosphate, and 0.02% magnesium sulfate, based on a previous study performed by Kuhlman, et al. [17].
Chemical mutagenesis
Inoculums preparation: A single colony of L. enzymogenes bacterial cells was transferred to 5 ml of nutrient broth and maintained at 33 ± 1 ºC with shaking at 180 rpm to reach an appropriate optical density (OD600nm), and then the inoculation was done by adding this sample to 50 ml of nutrient broth and incubated for an overnight.
NTG stock preparation
In the present study, NTG as a mutagenic substance was used to enhance the production of protease enzymes in L.enzymogenes. Initially, a stock solution of NTG was prepared by dissolving 0.01 g of NTG powder (TCI, Tokyo, Japan) in 1ml of dimethyl sulfoxide (DMSO, Merck, Germany). Various concentrations of NTG were made by diluting the primary stock in different amounts of 0.05M sodium phosphate buffer and then passing through a 0.22 µm syringe filter.
Mutagenesis procedure
Firstly, the OD600nm of the inoculated culture was measured and adjusted to 0.1 by 0.05M phosphate buffer saline (PBS). Then, the bacterial pellet was separated by centrifuging at 10,000 rpm for 5 min, washing 3 times with PBS, and then re-suspending in PBS. The suspension was treated with various concentrations of NTG (100, 150, and 200 µg/ml) for 20 and 40 min at 33 ± 1 ºC under shaking at 130 pm. After incubation times, the treated cells were separated by centrifugation at 10,000 rpm for 10 min and washed 3 times with PBS to discard NTG. Finally, 1 ml of fresh nutrient broth was added to treated bacterial cells and incubated at 33 ± 1 ºC under shaking at 130 rpm overnight. The procedure has been used was the modified methods of Lynn, et al. [18] and Zambare [19].
Assessing bacterial viability after NTG treatment
The bacterial colony-forming units (CFU) counting was exerted to evaluate the effect of NTG on the survival rate of bacteria after treatments. For this purpose, the OD600nm of wild-type (as control) and NTG-treated bacterial suspensions were adjusted to 1 and then serial dilutions (10-1, 10-2, 10-3, 10-1, and 10-5) were prepared in PBS. Then, 0.1 ml of diluted mutated bacteria were cultured on a nutrient agar medium and incubated at 33 ± 1 ºC for 48 h. For each sample, the dilutions which resulted in 30-300 numbers of colonies on the solid media were chosen for colony counting [20].
Screening methods
The initial screening stage is performed to select colonies with a clear hydrolysis zone of casein as substrate based on a previous study from Yokota, et al.[21], with some modifications. Single mutated clones were selected randomly from nutrient agar medium and cultured on skim milk agar plates (Merck, Germany). This medium was applied to isolate the mutants with proteolytic activity by exhibiting a zone of clearance around colonies. Then, in the secondary screening, the overnight cultures were prepared for each randomly selected NTG-treated bacterium and 20 µl of bacterial suspensions were transferred at the center of wells which were made in the 2.5% of skimmed milk agar. On each plate, one well devoted to wild-type bacterium and one for nutrient broth without bacteria, as a negative control. The plates were incubated at 33 ± 1 ºC for 48 h, and then the diameter of the clear zone around each mutated colony was measured and compared to the clear zone of the wild type. The procedure was done again in triplicates for those mutants that showed larger zones’ diameter than the wild-type strain. Then, the bacterial supernatants were examined and compared to the wild-type protease activity. The statistical analysis of casein hydrolysis zone increment of mutant isolates in contrast to the wild-type bacteria was performed using a one-way ANOVA test using Graph Pad Prism software, version 9.0.1. A p-value of less than 0.05 was considered a significant difference between the groups.
Proteolytic enzyme activity
Enzyme activity was further assessed by a modified version of Anson’s method [22] which measures the release of tyrosine from digested casein by proteases present in bacterial supernatant. For supernatant collection, an overnight culture of each selected mutant was added into a 250ml-Erlenmeyer flask containing 50ml of enzyme production medium and incubated under shaking for 2, 8, 10, 16, 24, and 48 h. After each incubation time, the cell-free supernatants were collected by centrifugation at 6000 rpm for 10 min. Then, 1 ml of supernatant was transferred into 2 ml casein solution (0.65% W/V in 0.05 M phosphate buffer, pH: 7) and the mixture was maintained for 30 min at 37 °C. The enzyme-substrate reaction was terminated by adding 2 ml of trichloroacetic acid (TCA, 110 mm, Merck, Germany) to the mixture and incubating for 30 min at 37 °C. In the following, the mixture was centrifuged at 6000 rpm for 10 min, the pellet was discarded, and 1 ml of supernatant was mixed with 5 ml of NaCO3 (0.5 M) and 1 ml of Folin–Ciocalteu reagent (0.5 M, Sigma, USA) and hold for 20 min at room temperature in a dark place. After the incubation period, OD was measured at 660 nm. The Enzyme unit was calculated by plotting a tyrosine standard curve (10–500 μg) and expressed as the amount of enzyme that released 1 μg.ml-1 of tyrosine/min according to the test condition (Zambare 2010; Anson 1938) [19,21].
Results
Mutagenesis and determination of survival rate
Random mutagenesis by NTG was employed to obtain mutant isolates with increased proteolytic activity. For this, wild-type strain was exposed to different concentrations of NTG at various time intervals. The viability percentage of mutants was measured by CFU counting for two concentrations (The highest and the lowest) of NTG (100 and 200 µg/ml) in 20 and 40 minutes exposure times. As shown in Table 1, the survival rate significantly decreased with an increasing dose of NTG, representing a dose-response pattern (Table 1). After 20 min exposure to 100 µg/ml of NTG, the survival rate decreased to 60.8 %, whereas the lowest survival rate was 31.1%, which was obtained after 40 min treatment with 200 µg/ml of NTG.
Screening of mutants on a skimmed milk agar plate
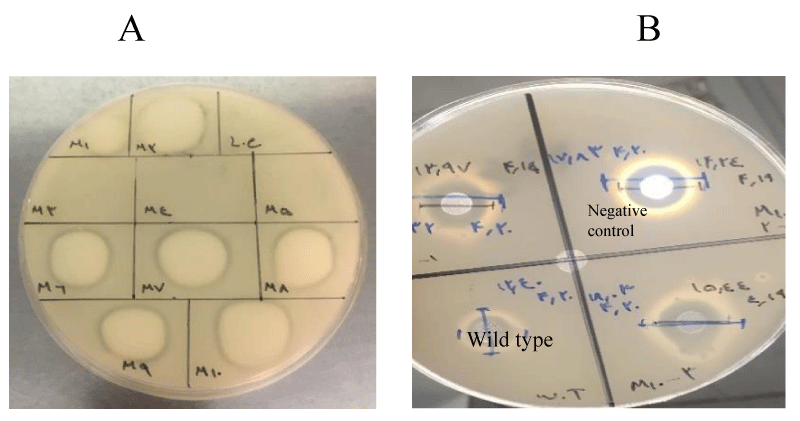
From NTG-treated bacteria, a number of colonies were randomly selected and streaked on 2.5% skimmed milk agar for primary screening. Based on the casein hydrolysis clearance zone, 30 mutants were selected for further investigation. As shown in figure 1, the proteolytic activity was disrupted in some of the mutants after NTG treatment which was excluded from the rest of the study (Figure 1A). Further screening was performed with the culture of 30 isolated mutants using the well diffusion method and carefully measuring the casein hydrolysis clearance zone (Figure 1B).
The secondary screening was performed on the mutated isolates which showed a larger zone of casein hydrolysis in contrast to the wild type. For this, supernatants were harvested from the logarithmic phase of the mutant and wild-type bacteria, and the casein hydrolysis zone diameter was measured and compared with the wild-type supernatant (all experiments were done as triplicates).
Among 30 examined strains, 11 mutated strains showed higher protease activity. However, based on the results demonstrated in Table 2, the clear zones around M4 (100 µg/ml for 20 min) and M10 (200 µg/ml for 40 min) mutants showed a statistically significant increment of casein hydrolysis zone in contrast to the wild type isolate; 22.07 and 21.45 mm, respectively vs. 17.04 in wile type. (One-way ANOVA test, p-value < 0.01; Table 2).
Determination of protease activity of selected mutants
During the secondary screening, both isolated mutants were grown in nutrient broth and the supernatants were collected during the logarithmic phase (OD600 = 0.4, and OD600 = 0.6, based on the results of a preliminary study the highest amount of protease was obtained during the logarithmic phase of L. enzymogenes growth) and they were cultured into the wells on 2.5% skimmed milk agar for 48 h. The comparison of hydrolysis zone diameters showed that there was no significant difference at OD600 = 0.4 between M4 mutants and wild type, whereas it was 1.17 times larger in M4 than wild types at OD600 = 0.6 (Table 3). However, the clearance zone for the M10 mutant was higher than the wild type in both ODs (1.25 times higher in OD600 = 0.4 and 1.10 times in OD600 = 0.6; Table 3).
To calculate the activity of the enzyme in units per milliliter, the supernatant of isolated mutants was examined using the Anson method and the results were calculated based on the standard tyrosine curve. Accordingly, the highest yield of protease was obtained in M10 mutants during the logarithmic phase compared to M4 and wild-type strains (246.235 and 270.71 U/ml in OD600 = 0.4 and OD600 = 0.6, respectively; Table 3). However, the proteolytic activity of M4 mutant was also higher than wild types (197.34 and 154.6 U/ml in OD600 = 0.4, and OD600 = 0.6, respectively; Table 3).
Discussion
Proteases are a large and extremely important group of enzymes in biology, medicine, and biotechnology. Microorganisms are a good source of enzymes with high diversity in biochemical properties as well as a better ability for genetic manipulation [1]. Lysobacter enzymogenes is a well-known gram-negative bacterium that produces different extracellular enzymes, especially proteases [23]. The most important of its enzymes is endopeptidase Lys-C, which is extensively used in biotechnological techniques such as primary-structure analysis, peptide mapping, in-gel digestion, and cleavage of fusion proteins in recent years [9]. However, the low yield of enzyme production from its source is the main concern for large-scale production. In this regard, modification of enzyme production pathway in such producer microorganisms is a crucial method to elevating production rate [24,25]. Random mutagenesis by chemical mutagens is one of the traditional approaches that is widely applied for strain improvement [26]. NTG is a very efficient mutagen from DNA alkylating agents which showed mutagenic effect by adding methyl to guanine and converting guanine to O6 methyl guanine [16].
Several previous studies exerted NTG mutagenesis to improve the production of various enzymes and metabolites in different microorganisms. Random mutagenesis by NTG on B. subtilis resulted in overproduction of Poly-γ-glutamic acid higher than the wild type in five selected isolates [27]. Based on such documentation, NTG mutagenesis was performed in the present study on L. enzymogenes to obtain mutants with increased proteolytic activity. To produce enough mutants with a high viability rate for screening, the fresh culture of L. enzymogenes was treated with different concentrations of NTG at various time intervals and the survival rate was evaluated by CFU counting. Based on the results, the survival percentage was decreased with elevating the NTG concentration and the exposure time. So that the lowest survival rate was 30.1% which obtain by treating bacterial suspension with 200 µg/ml of NTG after 40 min. The same trend of survivability reduction due to a higher concentration of the mutagenic agent has also been reported. In a study performed by Arshad, et al. [28], it was reported that treatment of Escherichia coli strains with NTG exhibited a dose and time-dependent pattern.
Another study conducted by Zambare in 2010 aimed to increase protease activity in Trichoderma reesei MTCC-3929 using a combination of UV and NTG mutagen. Mutant isolates were screened as protease producers based on the clearance zone diameter on the skimmed milk agar medium. Finally, a mutant called NTG-17 was found with a 2.6-fold increment of protease activity in contrast to the wild type. In the same manner, Zeng, et al. [29] reported that the isolate NCU116 produces approximately 31.9% fold more proteinase over the parental strain by combining ultraviolet irradiation and NTG treatment for mutagenesis.
In the present study, to isolate mutants with enhanced proteolytic activity, treated colonies were randomly streaked on 2.5% skim milk agar and all mutants with the ability to hydrolyze casein were selected. As a result of the screening procedure, 11 mutants that showed increased proteolytic activity in contrast to the wild type were selected. Among them, M4 (100 µg/ml for 20 min) and M10 (200 µg/ml for 40 min) mutants exhibited maximum proteolytic activity (Table 2). Since L. enzy mogenes normally released proteases during the logarithmic phase of the growth, supernatants of M4 and M10 were collected and analyzed during the logarithmic phase. Based on the casein hydrolysis zone, M10 mutant showed the highest protease activity which was 1.45- fold increment in OD600 = 0.4 in comparison with the wild type. Moreover, the unit of enzyme activity was qualified by a modified version of the Anson method which in the highest extracellular protease activity was obtained in M10 mutants during logarithmic phase compared to M4 and wild type strains which were 2.65 and 2.56 fold of wild type in OD600 = 0.4 and 0.6, respectively.
The earlier studies showed that the use of combinational mutagenesis by NTG and a physical mutagen made mutation more efficient. Accordingly, the combinational mutagenesis on the Bacillus pumilus strain created a mutant strain with increased protease activity (6000 U/ml in contrast to 1200 U/ml of wild type) [30]. Similarly, another study was conducted by Muhammad Nadeem, et al. [31]. They found that applying physical and chemical mutagenesis showed 1.4 fold higher protease activity in the pre-optimized growth medium than the parent strain. In our previous studies, physical mutagenesis using UV radiation and cold atmospheric plasma on the L. enzymogenes strain leads to isolating mutant strains with enhanced proteolytic activity with the highest ratio of 1.91 [32,33]. Therefore, it is predictable that the combination of UV and NTG could be also efficient for enhancing the proteolytic activity of L. enzymogenes which should be assessed in future studies.
Conclusion
Taken together, the results of the present study suggested that using NTG as a chemical mutagen can be a suitable method for generating mutants with higher proteolytic activity. However, the number of improved mutants and the amount of proteolytic activity increment in this method were relatively low; hence it seems that the use of NTG in combination with other mutagens would be a more potent way for the strain improvement.
This project was financially supported by the Iranian National Elites Foundation and Pasteur Institute of Iran.
- Razzaq A, Shamsi S, Ali A, Ali Q, Sajjad M, Malik A, Ashraf M. Microbial Proteases Applications. Front Bioeng Biotechnol. 2019 Jun 12;7:110. doi: 10.3389/fbioe.2019.00110. PMID: 31263696; PMCID: PMC6584820.
- Rani K, Rana R, Datt S (2012) Review on latest overview of proteases. Int J Curr Life Sci 1:12-18.
- Kasana RC, Salwan R, Yadav SK. Microbial proteases: detection, production, and genetic improvement. Crit Rev Microbiol. 2011 Aug;37(3):262-76. doi: 10.3109/1040841X.2011.577029. Epub 2011 May 21. PMID: 21599542.
- Banerjee G, Ray AK. Impact of microbial proteases on biotechnological industries. Biotechnol Genet Eng Rev. 2017 Oct;33(2):119-143. doi: 10.1080/02648725.2017.1408256. Epub 2017 Dec 5. PMID: 29205093.
- Xu G, Zhao Y, Du L, Qian G, Liu F. Hfq regulates antibacterial antibiotic biosynthesis and extracellular lytic-enzyme production in Lysobacter enzymogenes OH11. Microb Biotechnol. 2015 May;8(3):499-509. doi: 10.1111/1751-7915.12246. Epub 2015 Feb 13. PMID: 25683974; PMCID: PMC4408182.
- Nijhuis EH, Pastoor R, Postma J. Specific detection of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8 with TaqMan PCR. J Appl Microbiol. 2010 Apr;108(4):1155-66. doi: 10.1111/j.1365-2672.2009.04519.x. Epub 2009 Aug 11. PMID: 197322
- Gómez Expósito R, Postma J, Raaijmakers JM, De Bruijn I. Diversity and Activity of Lysobacter Species from Disease Suppressive Soils. Front Microbiol. 2015 Nov 16;6:1243. doi: 10.3389/fmicb.2015.01243. PMID: 26635735; PMCID: PMC4644931.
- Xie Y, Wright S, Shen Y, Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012 Nov;29(11):1277-87. doi: 10.1039/c2np20064c. PMID: 22898908; PMCID: PMC3468324.
- Chohnan S, Nonaka J, Teramoto K, Taniguchi K, Kameda Y, Tamura H, Kurusu Y, Norioka S, Masaki T, Sakiyama F. Lysobacter strain with high lysyl endopeptidase production. FEMS Microbiol Lett. 2002 Jul 16;213(1):13-20. doi: 10.1111/j.1574-6968.2002.tb11279.x. PMID: 12127482.
- Stressler T, Eisele T, Meyer S, Wangler J, Hug T, Lutz-Wahl S, Fischer L. Heterologous expression and pro-peptide supported refolding of the high specific endopeptidase Lys-C. Protein Expr Purif. 2016 Feb;118:31-8. doi: 10.1016/j.pep.2015.09.024. Epub 2015 Sep 30. PMID: 26431800.
- Wu Z, Huang J, Huang J, Li Q, Zhang X. Lys-C/Arg-C, a More Specific and Efficient Digestion Approach for Proteomics Studies. Anal Chem. 2018 Aug 21;90(16):9700-9707. doi: 10.1021/acs.analchem.8b02448. Epub 2018 Aug 1. PMID: 30024741.
- Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000 Sep;54(3):287-301. doi: 10.1007/s002530000403. PMID: 11030563.
- Sturtevant J. Site-Directed and Random Insertional Mutagenesis in Medically Important Fungi. In: Genetic Manipulation of DNA and Protein-Examples from Current Research, first ed.; Figurski, D. Intech Open: Columbia University, United States of America. 2013. https://doi.org/10.5772/47799.
- Korbekandi H, Darkhal P, Hojati Z, Abedi D, Hamedi J, Pourhosein M. Overproduction of Clavulanic Acid by UV Mutagenesis of Streptomyces clavuligerus. Iran J Pharm Res. 2010 Spring;9(2):177-81. PMID: 24363725; PMCID: PMC3862066.
- Tasneem M, Khan A, Ashraf H, Haq I. Xylanase Biosynthesis by chemically mutated strain of Aspergillus niger. J Food Technol. 2003; 1(4):178-81.
- Huovinen T. Methods of genetic diversity creation and functional display for directed evolution experiments [Dissertation on the Internet]. University of Turku. 2014. : https://www.utupub.fi/handle/10024/94147.
- Kuhlman PA, Chen R, Alcantara J, Szarka S. Rapid purification of Lys-C from Lysobacter enzymogenes cultures: A sequential chromatography technique. BioProcess Int. 2009; 7:28-38.
- Lynn TM, Yu SS, Kyaw EP, Latt ZK. Enhancement of phosphate solubilizing activity of Enterobacter clocae by NTG mutagenesis. Sky J Soil Sci Environ Manag. 2014; 2:102-10.
- Zambare V. Strain improvement of alkaline protease from Trichoderma reesei MTCC-3929 by physical and chemical mutagen IIOAB J. 2010; 1: 25-28.
- Li HG, Luo W, Gu QY, Wang Q, Hu WJ, Yu XB. Acetone, butanol, and ethanol production from cane molasses using Clostridium beijerinckii mutant obtained by combined low-energy ion beam implantation and N-methyl-N-nitro-N-nitrosoguanidine induction. Bioresour Technol. 2013 Jun;137:254-60. doi: 10.1016/j.biortech.2013.03.084. Epub 2013 Mar 21. PMID: 23587827.
- Yokota K, Furusawa N, Abe T, Takenaka S. Application of casein agar plate method for the determination of protease activity. Eisei Kagaku. 1988; 34:241-247.
- Anson ML. THE ESTIMATION OF PEPSIN, TRYPSIN, PAPAIN, AND CATHEPSIN WITH HEMOGLOBIN. J Gen Physiol. 1938 Sep 20;22(1):79-89. doi: 10.1085/jgp.22.1.79. PMID: 19873094; PMCID: PMC2213732.
- Yin H. Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes [Dissertations and Theses in Biological Sciences]. Lincoln: University of Nebraska. 2010. https://digitalcommons.unl.edu/bioscidiss/15.
- Vittaladevaram V. Fermentative Production of Microbial Enzymes and their Applications: Present status and future prospects. J Appl Biol Biotechnol. 2017; 5(04):90-94. https://doi.org/10.7324/JABB.2017.50414
- Gurung N, Ray S, Bose S, Rai V. A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. Biomed Res Int. 2013;2013:329121. doi: 10.1155/2013/329121. Epub 2013 Sep 11. PMID: 24106701; PMCID: PMC3784079.
- Heerd D, Tari C, Fernández-Lahore M. Microbial strain improvement for enhanced polygalacturonase production by Aspergillus sojae. Appl Microbiol Biotechnol. 2014 Sep;98(17):7471-81. doi: 10.1007/s00253-014-5657-z. Epub 2014 Apr 4. PMID: 24695827.
- Mahidsanan, T, Gasaluck, P. Improvement of poly-γ-glutamic acid (PGA) producing Bacillus subtilis SBMYP- 1 by N-methyl-N'-nitro-N-nitrosoguanidine (NTG) mutagenesis. Int Food Res. 2016; J 23(2):751-755.
- Arshad R, Farooq S, Ali SS. Effect of mutations induced by N-methyl-N′-nitro-N-nitrosoguanidine on expression of penicillin G acylase and β-lactamase in wild-type Escherichia coli strains. Ann Microbiol 60:645-652. https://doi.org/10.1007/s13213-010-0104-6.
- Zeng C, Zhao R, Ma M, Zeng Z, Gong D. Mutagenesis and characterization of a Bacillus amyloliquefaciens strain for Cinnamomum camphora seed kernel oil extraction by aqueous enzymatic method. AMB Express. 2017 Dec;7(1):154. doi: 10.1186/s13568-017-0451-9. Epub 2017 Jul 17. PMID: 28724263; PMCID: PMC5514006.
- Wang HY, Liu DM, Liu Y, Cheng CF, Ma QY, Huang Q, Zhang YZ. Screening and mutagenesis of a novel Bacillus pumilus strain producing alkaline protease for dehairing. Lett Appl Microbiol. 2007 Jan;44(1):1-6. doi: 10.1111/j.1472-765X.2006.02039.x. PMID: 17209806.
- Nadeem M, Qazi JI, Baig S. Enhanced Production of Alkaline Protease by a Mutant of Bacillus licheniformis N-2 for Dehairing. Braz Arch Biol Technol. 2010. 53(5):1015-1025.
- Kalahroudi RJ, Valizadeh V, Atyabi SM, Keramati M, Cohan RA, Aghai A, Norouzian D. Increment in protease activity of Lysobacter enzymogenes strain by ultra violet radiation. Iran J Microbiol. 2020 Dec;12(6):601-606. doi: 10.18502/ijm.v12i6.5035. PMID: 33613915; PMCID: PMC7884282.
- Tabar FF, Valizadeh V, Keramati M, Davoudi M, Molasalehi S, Fakhabi NS, Atyabi SM, Cohan RA, Norouzian D. Enhancing proteolytic activity of Lysobacter enzymogenes using cold atmospheric plasma. Arch Microbiol. 2022 May 20;204(6):343. doi: 10.1007/s00203-022-02936-4. PMID: 35596084.

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
