Journal of Clinical Microbiology and Biochemical Technology
Phenotypic characterization of bacterial isolates from marine waters and plastisphere communities of the Ross Sea (Antarctica)
Gabriella Caruso1*, Ombretta Dell’Acqua2, Rosalba Caruso3 and Maurizio Azzaro4
2Department of Earth, Environment and Life Sciences, University of Genoa, Genoa, Italy
3Hospital Agency A.O.U. “G. Martino”, Messina, Italy
4CNR-ISP, Messina, Italy
Cite this as
Caruso G, Dell’Acqua O, Caruso R, Azzaro M (2022) Phenotypic characterization of bacterial isolates from marine waters and plastisphere communities of the Ross Sea (Antarctica). J Clin Microbiol Biochem Technol 8(1): 001-009. DOI: 10.17352/jcmbt.000048Copyright
© 2022 Caruso G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: In aquatic environments, microbial biofilms are hot spots of microbial diversity, as well as a substrate for larval settlement of many invertebrate species. Examining the functional diversity of microorganisms in polar regions is a new and still unknown field of aquatic microbiology, that is attracting increasing interest for its significance in both scientific research and resources exploitation.
The context and purpose of the study: In the framework of research aimed at studying microbial colonization occurring at Terra Nova Bay, culturable heterotrophic bacteria were quantified and characterized phenotypically at two study areas (Road Bay and Tethys Bay) with different environmental characteristics: the first one was close to Mario Zucchelli research station and impacted by anthropogenic pressure due to sewage wastes, while the second was exposed to salinity gradients due to glacier melting. In this context, artificial structures hosting plastic (polyvinyl chloride, PVC and polyethylene, PE) panels were deployed at -5 and -20m depths at each of the stations (one Impact and one Control) chosen per each study area. Water samples were contextually collected at the time of recovery of the plastic panels, namely after 12 months of deployment; at Road Bay, at the -5 m depth, a short-term experiment was also performed, with the recovery of the panels after 2.5 and 9 months of deployment. Bacterial strains were isolated in axenic culture both from water and biofilm matrices, and examined for their main phenotypical traits and functional diversity, through Gram staining, oxidase production, glucose fermentation and screened for enzymatic activity profiles (proteolytic, glycolytic and phosphatasic activities) using specific fluorogenic substrates.
Results: Most of the bacterial isolates were Gram-negative, oxidase-positive and glucose-fermenting strains. Higher enzyme diversification was found in Road Bay at the site impacted by the sewage wastes from the research station, compared to the control site. A higher abundance of heterotrophic bacteria was detected on PVC compared to PE. Several pigmented strains, ascribable to Flavobacterium sp., were isolated from biofilm with respect to the ones isolated from the pelagic environment.
Main findings: The screening of bacterial isolates for the production of hydrolytic enzymes revealed that proteolytic and phosphatase activities were a common metabolic trait of both water and biofilm-derived isolated. Microbes inhabiting Antarctica represent a very important and not completely known source of bioactive molecules with possibly multiple applications, from biomedicine to pharmaceutical, cosmetics, biodegradation, and so on.
Conclusions: This is the first contribution to characterize phenotypically the bacteria colonizing the Ross Sea seabed in comparison with pelagic heterotrophic bacteria and to explore their metabolic abilities and the biotechnological potential of these microorganisms. The preliminary data regarding the metabolic profiles and composition of the microbial community have highlighted the plasticity of the microbial community, whose metabolism is modulated by the organic matter supplies found near the Mario Zucchelli Station.
Brief summary: Heterotrophic bacteria in the waters and in the biofilm covering the surface of settlement panels in plastics deployed at sea for different time periods were studied in two bays of the Ross Sea, Road Bay and Tethys Bay. The main phenotypical characteristics of the bacterial isolates were examined. The production of hydrolytic enzymes makes microbial biofilms interesting sources of molecules with a potential applicative interest.
Any potential implications: Bioprospecting research on polar microorganisms represents an attractive field of microbiology that is supported by increasing attention towards the discovery of hydrolytic enzymes as new active compounds having unexplored properties and applications.
Introduction
In aquatic ecosystems, biofilms cover the surface of all submerged substrates [1]. Marine biofilms are complex systems consisting of microbes embedded in a matrix of extracellular polymers [2]. Microbial biofilms represent hot spots of microbial diversity and a potential source of secondary metabolites; acting like a second skin on the organisms [3], they are also suitable substrates for the larval settlement of many invertebrate species [4]. Microbial communities (bacteria and microalgae) within biofilm follow distinct succession patterns and fill specific functional niches; they are able to structurally self-organize in response to external conditions and to the activities of the various biofilm members. Biofilm-associated bacteria and microalgae also contribute to the overall functioning of aquatic ecosystems, by regulation of key biological processes (i.e. degradation of organic matter and environmental pollutants, photosynthesis, nitrogen fixation) [5,6].
Despite the increasing interest in microbial biofilms, in polar regions such as Antarctic marine environments, there is still a paucity of knowledge on community composition in most marine biofilms and their responses to natural/anthropic changes [3, 4, 7]. Particularly, the microbial colonization in the Ross Sea is largely unexplored and increasing human pressure makes a clearer understanding of microbial community dynamics necessary [6]. The interest in microbial colonization is also related to the potential threat posed by marine litter and particularly plastic pollution that has come to an emerging field of research for its eventual detrimental consequences for human and animal health [6] as well as for the overall ecosystem health. Plastics are a unique habitat to host and spread across the oceans diverse microbial species attached to their surface, creating the so-called “plastisphere”, according to the name coined by Zettler, et al. [8]. The plastisphere community composition seems to be affected by the chemical nature of plastic substrates of different compositions [9].
For long times, Antarctic regions have been considered pristine environments, but recent evidence of human footprints on these vulnerable ecosystems has been provided [10-13]. Furthermore, the microorganisms that survive and grow in extremely cold conditions such as those typical of polar regions have adopted a variety of adaptive strategies that allow them to maintain their activity and metabolic function despite the challenging conditions. These microorganisms are expected to possess properties of interest in several fields of applications, such as biomedical, cosmetical, bioremediation, and so on [14-15].
In the light of the above-reported considerations, funded by the Italian National Research Program in Antarctica (PNRA), research has recently been undertaken in the Ross Sea in order to follow the process of microbial colonization in relation to the further settlement of benthic communities and to assess microbial abundance and metabolism in two coastal sites differently exposed to natural and anthropogenic forcings.
The specific objectives of the present study were: i) to assess quantitatively the heterotrophic bacteria present in two Antarctic sites exposed to anthropic and natural perturbations and ii) to study the phenotypical characteristics of bacterial isolates in order to get insights into their composition and functional diversity.
Materials and methods
Study area and experimental design
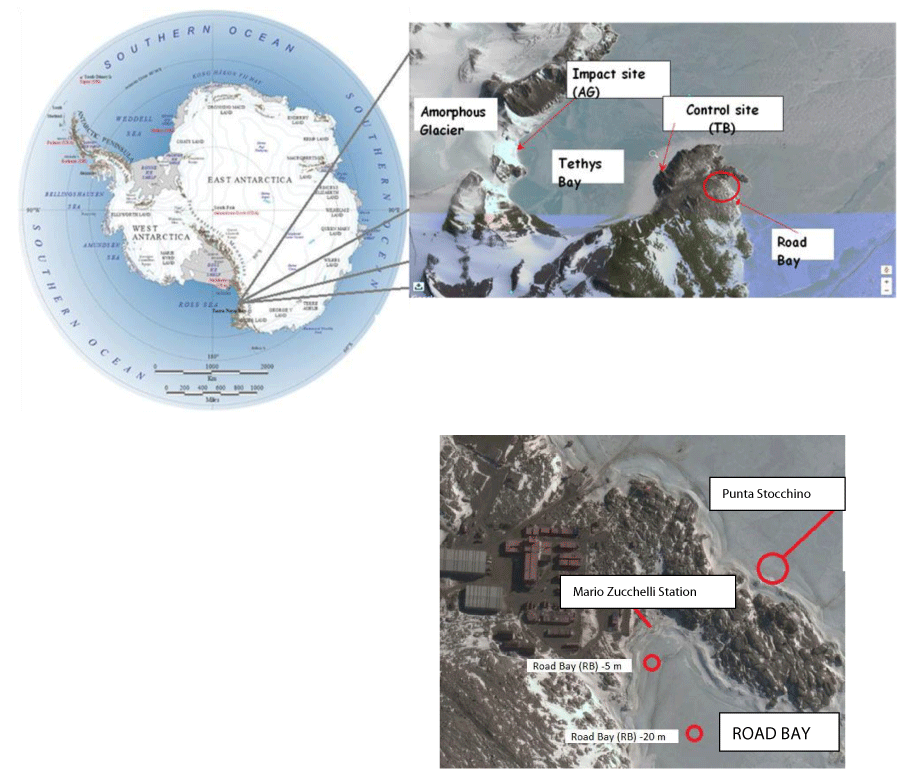
The research was performed during the XXXIII and XXXIV Italian Antarctic expeditions, covering two consecutive austral summer periods. Two different areas were chosen for this study (Figure 1).
- Road Bay, a small bay close to the Mario Zucchelli Research Station, where the Research Station sewage is located and that represented a site exposed to anthropic impact.
- Tethys Bay, a large bay 2 km North of Road Bay, is inner and isolated from Road Bay. Herein a large glacier (Amorphous Glacier) approaches the shore and a salinity gradient was expected to be present; this site allowed to simulate potential effects caused by global warming.
At each of the study areas an impact site and a control site were set up, and precisely Road Bay (RB, Impact) and Punta Stocchino (PTS, Control) within the Road Bay area; Amorphous Glacier (AG, Impact) and Tethys Bay (TB, Control) within the Tethys Bay area. At all these stations artificial structures with attached plastic panels were deployed at depths of -5 and - 20 meters. The geographical coordinates of the study sites were: RB (Latitude 74°41,743’ S Longitude 164°07,125’ E), PTS (Latitude 74°41,651’ S Longitude 164°07,303’ E), TB (Latitude 74°41,417’ S Longitude 164°06,303’ E) ; AG (Latitude 74°41,234’ S Longitude 164°02,135’ E).
Settlement panels for microbial colonization
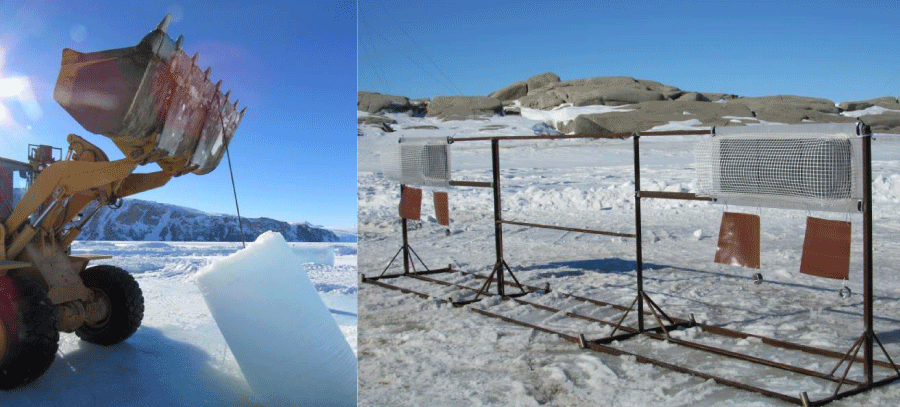
Polycarbonate Vinyl Chloride (PVC) and Polyethylene (PE) panels (approximately 15 cm x 15 cm) were used as artificial substrates for the study of microbial colonization. The plastic panels - in a number of 3 PVC and 2 PE plates - were fixed into a frame in inox (Figure 2); in the pack ice large holes were produced, through which each structure was submerged and moored with hooks at established depths (-5 and -20 m).
Two experiments (short-term and long-term experiments) were performed. For the long-term experiment, the panels were fixed at the arrival in Antarctica (November 2017, during the first expedition) and removed after one year (in November 2018) using divers who plunged from holes made by ENEA logistics in the pack-ice. In the short-term experiment, performed in Road Bay at -5 m depth only, all the panels were removed after 2.5 months of immersion and replaced with new ones, which were collected at the beginning of the successive year, therefore after 9 months.
All the recovered panels were assessed for the amount of biofilm, and the abundance and functional diversity of bacterial biofilm components. At the same time of recovery of the plastic panels (i.e. after 12 months of immersion), water samples were collected and temperature, salinity, dissolved oxygen, pH and fluorescence measurements were taken along the water column too.
Bacteriological analysis: Cultivation procedure and Isolation of bacterial strains
The viable heterotrophic bacteria were estimated by inoculation of serial dilutions of water and biofilm samples on Marine Agar 2216 plates (Difco) incubated at +5°C for 7-21 days and the grown colony forming units (CFU) were counted. Colonies grown on Marine Agar were isolated; about 30 colonies were isolated from each sample, although some strains lost their viability during successive replications.
Phenotypic characterization
The phenotypic characterization of the isolated strains was performed according to the standard procedures in use at the CNR- ISP laboratory (Messina, Italy). All the bacterial isolates were screened for Gram staining, oxidase production, colony pigmentation, glucose fermentation and production of proteolytic, glycolytic and phosphatase enzymes as an expression of potential virulence factors [16]. The morphotype (i.e. cocci, rods, pleomorphs) was also recorded.
Gram staining
A Gram staining kit (Oxoid), composed of crystal violet, decolorizer, iodine solution and safranin was used to distinguish the Gram-negative strains (visible with a pink coloration) from the positive ones (colored in violet).
Oxydase test
The oxidase test is a biochemical reaction commonly used to search for the presence of cytochrome oxidase. When a microorganism contains the cytochrome oxidase enzyme, the reduced colorless Kovacs reagent (1% tetramethyl-p-phenylenediamine dihydrochloride) is converted into an oxidized colored product [17].
Glucose fermentation
All the strains were screened for their ability to ferment glucose on Triple Sugar Iron agar (Oxoid) tubes; the tubes were scored as positive (glucose-fermenting strains) or negative (glucose oxidizing strains or strains showing no reaction). The production of gas or hydrogen sulphide was also recorded as distinctive characters for the presumptive identification of each bacterial strain (see below).
Enzymatic profiles
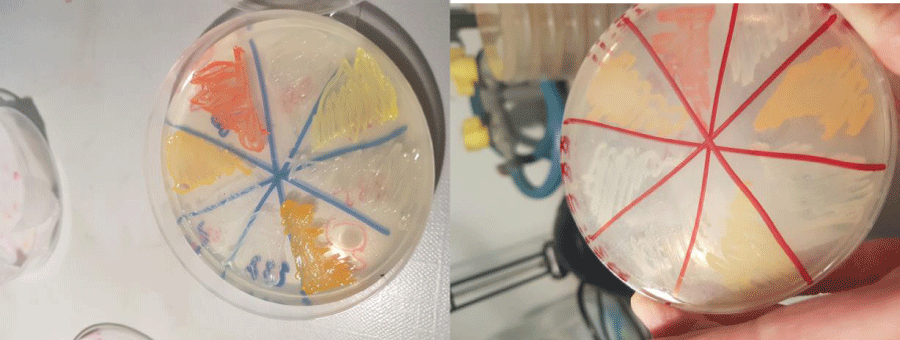
To test the bacterial ability to synthesize proteolytic (leucine aminopeptidase, LAP), glycolytic (alpha- and beta-glucosidase, GLU), phosphatasic (alkaline phosphatase, AP) enzymes all the strains in axenic culture were inoculated by replica plating on the surface of four Marine agar dishes, one for each enzyme to be assayed. Only one strain was inoculated per each Petri dish, in order to avoid potential interferences in the interpretation of the results [18]. After incubation for 10 days at +5°C, the plates were covered with a paper filter dish soaked by a 0.1 mM solution of each fluorogenic substrate [leucine-7 amido-4-methyl-coumarin, 4-Methlumbelliferyl (MUF)-a-d-glucoside, 4-MUF-b-d-glucoside, MUF-phosphate] specific for LAP, alpha-GLU, beta-GLU and AP, according to the Kim and Hoppe’s method [19]. The substrate hydrolysis was visible, after 3 min, as a fluorescent spot, around the bacterial colony under UV light of a Wood lamp.
The enzymatic profiles of bacteria isolated from water and biofilm at Road Bay (RB) and Punta Stocchino (PTS) were represented as heatmaps, that give a visual representation of the functional diversity as a panel of colors. Heatmaps were generated using the PAST software (version 3.14) [20].
Presumptive identification
According to their main morphological and physiological characteristics, bacterial isolates were identified at least at a genus level, using Oliver’s identification scheme [21].
Data representation
The phenotypical traits of bacterial isolates were represented as pie charts, that showed per each trait (Gram staining, morphotype, pigmentation, oxidase production, glucose fermentation) the percentage of positive strains with respect to the total number of isolates.
Results
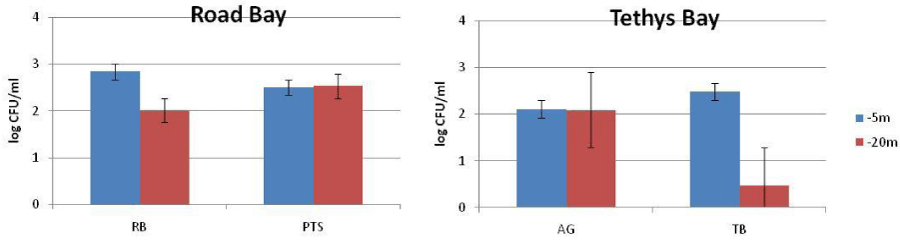
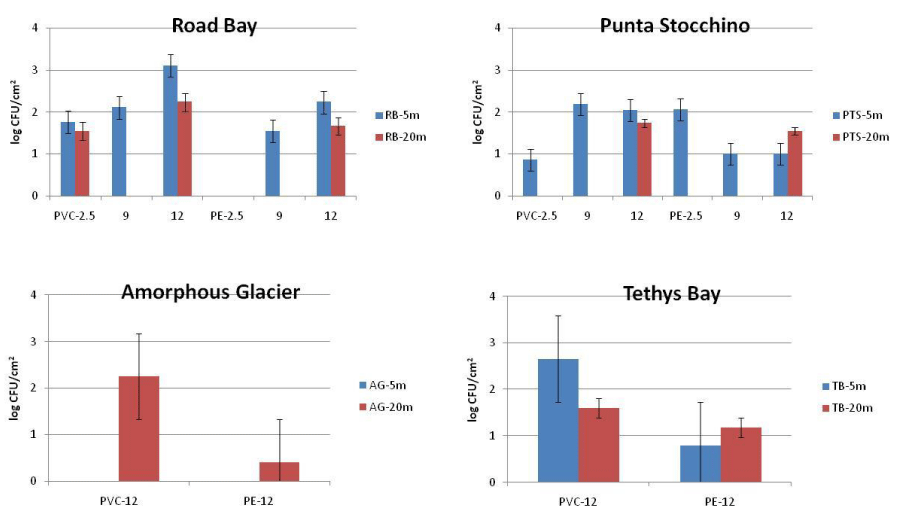
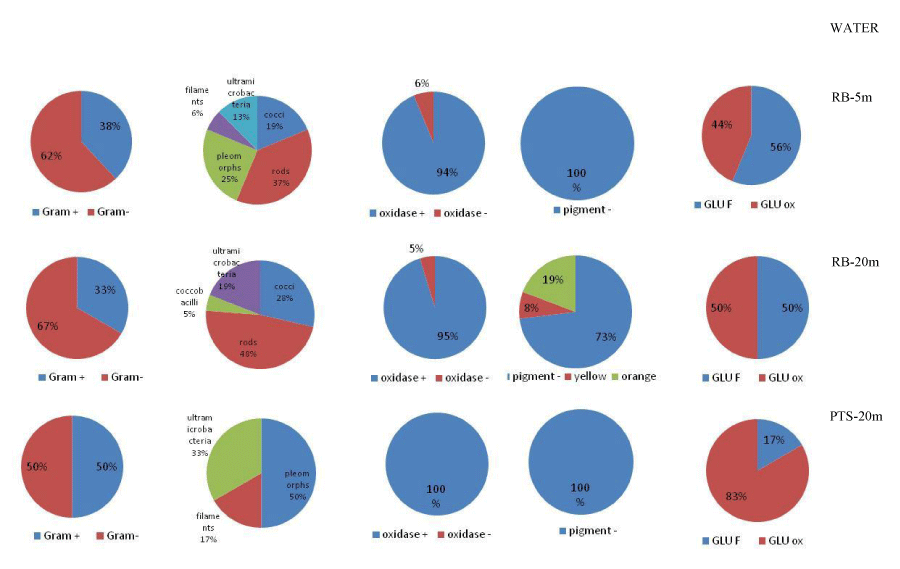
The abundance of culturable heterotrophic bacteria in water ranged from 1.03 x 102 to 6.87 x 102 CFU/ml in Road Bay and from 3 CFU/ml to 3.03 x102 CFU/ml in Tethys Bay. Within the biofilm matrix developed in Road Bay, their abundances were comprised between 3.5 x101 and 1.27 x 103 CFU/cm2 and between 3.5 x 101 and 1.71 x 102 CFU/cm2 for PVC and PE, respectively. In Tethys Bay, culturable heterotrophic bacteria varied from 4 x 101 to 4.52 x 102 CFU/cm2 and from 3 to 1.5 x 101 CFU/cm2 for PVC and PE, respectively. Their distribution in the pelagic domain at the time of the substrate recovery and after 2.5, 9 and 12 months of substrate immersion for the biofilm is shown in Figures 3,4 for water and biofilm respectively. Both in water and biofilm, the heterotrophic bacteria were more abundant at the Impact than at the Control stations.
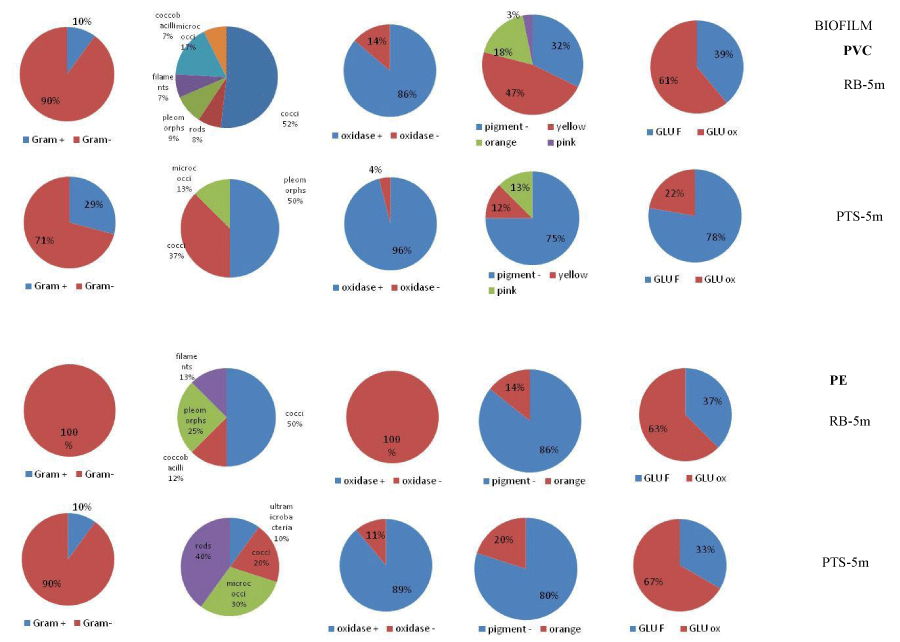
Several strains isolated from biofilm were pigmented (Figure 5), while pigmented strains were isolated from water only rarely.
Figures 6,7 summarize the main phenotypic characteristics shown by the bacterial strains isolated from the water and biofilm samples assayed in this study, respectively. In waters, Gram-negative strains were predominant (over 60% of the total); they were mostly oxidase-positive, not pigmented strains (excepting some yellow-orange pigmented strains found at RB-20m) and glucose-fermenting strains predominated at RB-5m, while glucose oxidizing strains prevailed at PTS-20m. Morphotypes were more diversified at RB-5m than at other sites. Bacterial strains isolated from the biofilm grown on PVC panels were mostly Gram-negative (over 70% of the total), oxidase-positive and glucose-fermenting strains; at the impact RB site they showed a diversified shape and pigmentation compared to the isolates from the control PTS site. Bacterial strains isolated from the biofilm grown on PE panels were characterized by a low degree of diversification compared to the analogous isolated from PVC. All strains were Gram-negative and oxidase-negative at the RB site.
The phenotypical differences found between the PVC- and PE-associated bacteria indicated that the two plastic materials represented different substrates for the attached microorganisms; moreover, the plastisphere was composed of microbial species well adapted to colonize these artificial substrates through diversified metabolic patterns.
In the presumptive identification based on the traditional biochemical methods, each isolate was assigned to one of the following main genera:
- -Pseudomonas: Gram-negative, glucose nonfermenting, oxidase-positive or -negative rods;
- -Vibrio/Aeromonas Photobacterium: Gram-negative, glucose-fermenting and oxidase-positive rods;
- -Flavobacterium: Gram-negative, pigmented, oxidase-positive or negative rods;
- -Bacillus: Gram-positive rods, with spores;
- -Corynebacterium: Gram-positive rods, without spores;
- -Micrococcus: Gram-positive, glucose-oxidative, clumps-shaped cocci.
According to these identification criteria, most of the bacterial isolates were assigned to Vibrio/Aeromonas/Photobacterium genera; the pigmented strains were classified as belonging to the Flavobacterium genus, while glucose nonfermenting strains were identified as Pseudomonas sp.
Some strains were assigned to Micrococcus sp.
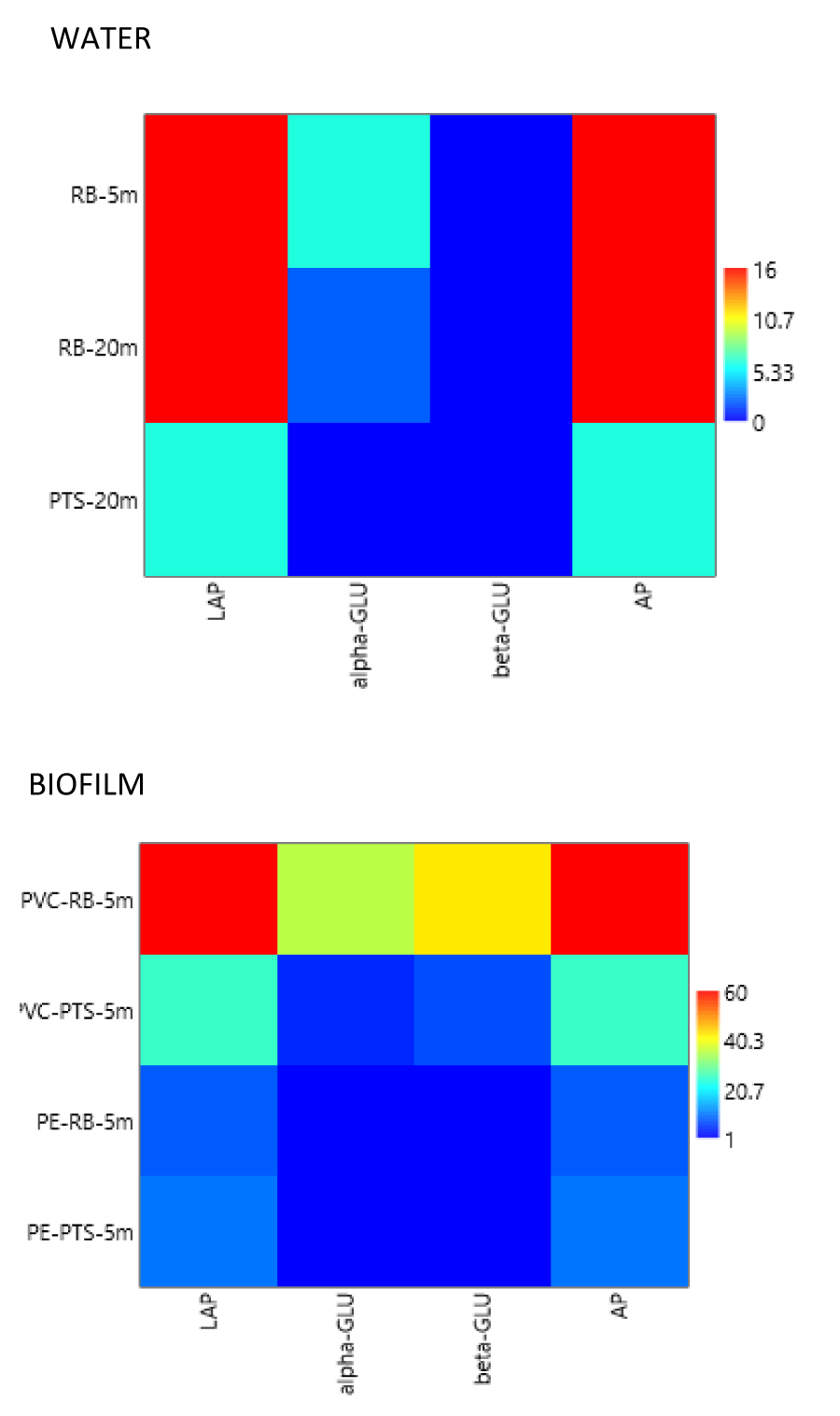
Enzymatic profiles, shown in Figure 8, indicated that the majority of the isolates were active producers of LAP and AP; another represented enzyme was beta-GLU, while a low number of strains produced alpha-GLU. Biofilm isolates showed higher enzymatic levels than water isolates.
Discussion
In the Antarctic environment, studies on the structure and function of the microbial communities that make up biofilms are still limited [4,7]. This study is the first to contribute to the knowledge of culturable bacteria belonging to microbial biofilm in the Ross Sea. In natural aquatic ecosystems, the community composition of microbial biofilms is widely variable; shifts in response to environmental or anthropogenic drivers are known. Except for a study at McMurdo Station [4] , to our knowledge, the marine microbial environment in Antarctica is largely unexplored, particularly regarding the response of biota to natural/anthropogenic pressures. Extreme environments such as polar regions are also among the most interesting study areas for the discovery of new microbial species, not yet cultured and explored. In this study, the high abundance of heterotrophic bacteria found at the Impact stations compared to the Control ones was likely caused by the water enrichment due to the Zucchelli wastewater depuration plant in the Road Bay area or to the release of detritus in proximity of the glacier in the Tethys Bay area. The most common bacteria isolated from water and biofilm were Gram-negative rods. Most of the isolates were assigned to the genera Vibrio/Aeromonas/Photobacterium; moreover, a frequently recovered genus was Pseudomonas, which is characterized by high metabolic versatility [18].
Bioprospecting is a quite recent field that is in progress; several studies have been previously performed on bacterial strains isolated from polar environments to explore their biotechnological potential (see [15] for a review). The ability to produce hydrolytic enzymes was observed in a wide number of bacterial strains isolated from water and biofilm samples from the Ross Sea. As it is known, the culturable fraction represents the viable fraction associated with organic polymers and is also equipped with a wide enzyme spectrum [18]. The enzymes assayed in the Ross Sea were among those related to the decomposition of common organic substrates. A high functional diversity of culturable bacteria was observed in Potter Cove, South Shetland Islands, Antarctica, where bacterial strains produced several hydrolytic enzymes, that represented a promising source of biomolecules [22]. Among 189 isolated bacteria, the strains able to produce amylases, pectinases, cellulases, CM-cellulases and proteases accounted for percentages of 9.5%, 22.8, 14.8%, 25.4%, and 44.4% of the total of strains. At least one extracellular enzyme was produced by more than 25% of the isolates, some of which were found to be able to produce up to six of the assayed enzymatic activities.
A broad range of cold-active hydrolytic enzymes was produced at a cultivation temperature of 4°C by most of the bacterial strains isolated from Arctic sea ice and seawater (Spitzbergen) samples [23]. The strains that were able to degrade proteins (using skim milk and casein as substrates), lipids (olive oil), and polysaccharides (starch and pectin) represented a percentage of 56, 31, and 21% of the total of isolated strains, respectively. Conversely, Fenice, et al. [24] found that a low eco- nutritional versatility in fungal isolates from different sites on Victoria Land (continental Antarctica). Glucose oxidase, protease and DNAase were almost low or absent in thirty-three fungal strains, while amylase and phosphatase activities were common and lipases were generally present in high quantities in almost all the strains.
Another interesting feature of the phenotypical study performed in the Ross Sea was the wide number of pigmented strains isolated from the biofilm The production of pigment has been suggested as a strategy to overcome oxidative stress [25,26]; another hypothesis is that pigment is a trait common to virulent strains [27].
Conclusion
This study was a contribution to current knowledge on the abundance and functional diversity of the culturable fraction of the heterotrophic bacterial biofilm community and to the discovery of new species and microbial products, as well as to clarify diversity-function relationships and interactions between the different bacterial groups in the Ross Sea benthic domain. New insights on plastic-associated bacterial assemblage adapted to live in this extreme environment were also provided.
This study was funded by the Italian PNRA project “Microbial colonization of benthic Antarctic environments: response of microbial abundances, diversity, activities and larval settlement to natural or anthropogenic disturbances and search for secondary metabolites” (ANT-Biofilm, PNRA16_00105). Thanks are due to the Barbier Group (France) for providing the polyethylene panels used in this study and to the ENEA staff who supported with logistical activities.
- Dang H, Lovell CR (2015) Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol Mol Biol Rev 80: 91-138. Link: https://pubmed.ncbi.nlm.nih.gov/26700108/
- Salta M, Wharton JA, Blache Y, Stokes KR, Briand JF (2013) Marine biofilms on artificial surfaces: structure and dynamics. Environ Microbiol 15: 2879-2893. Link: https://bit.ly/3K5hlTy
- Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F (2012) The second skin: ecological role of epibiotic biofilms on marine organisms. Front Microbiol 3: 292. Link: https://bit.ly/3zPPguG
- Webster NS, Negri AP (2006) Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ Microbiol 8: 1177–1190. Link: https://bit.ly/3nitbzS
- Anderson OR (2016) Marine and estuarine natural microbial biofilms: Ecological and biogeochemical dimensions. AIMS Microbiol 2: 304-331. Link: https://bit.ly/3K3n5gj
- Caruso G (2020) Microbial Colonization in Marine Environments: Overview of Current Knowledge and Emerging Research Topics. J Mar Sci Eng 8: 78. Link: https://bit.ly/3qiOfIt
- Lee YM, Cho KH, Hwang K, Kim EH, Kim M, et al. (2016) Succession of bacterial community structure during the early stage of biofilm development in the Antarctic marine environment. Korean J Microbiol 52: 49-58. Link: https://bit.ly/3r8NUXR
- Zettler ER, Mincer TJ, Amaral-Zettler LA (2013) Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ Sci Technol 47: 7137-7146. Link: https://bit.ly/3zOsseT
- Amaral-Zettler LA, Zettler ER, Slikas B, Boyd GD, Melvin DW, et al. (2015) The Biogeography of the Plastisphere: Implications for Policy. Front Ecol Environ 13: 541-546. Link: https://bit.ly/3K134XZ
- Lo Giudice A, Caruso G, Rizzo C, Papale M, Azzaro M (2019) Bacterial communities versus anthropogenic disturbances in the Antarctic coastal marine environment. Environmental Sustainability 2: 297-310. Link: https://bit.ly/3qjbjql
- Laganà P, Caruso G, Corsi I, Bergami E, Venuti V, et al. (2019) Do plastics serve as a possible vector for the spread of antibiotic resistance? First insights from bacteria associated to a polystyrene piece from King George Island (Antarctica). Int J Hygiene Environ Health 222: 89-100. Link: https://bit.ly/31PCnUV
- Cappello S, Caruso G, Bergami E, Macrì A, Venuti V, et al. (2021) New insights into the structure and function of the prokaryotic communities colonizing plastic debris collected in King George Island (Antarctica): preliminary observations from two plastic fragments. J Hazard Mater 414: 125586. Link: https://bit.ly/3ql8t4s
- Caruso G, Bergami E, Singh N, Corsi I (2012) Plastic human footprint in Antarctic waters and biota. Water Biology and Security.
- de Pascale D, De Santi C, Fu J, Landfald B (2012) The microbial diversity of Polar environments is a fertile ground for bioprospecting. Mar Genomics 8: 15-22. Link: https://bit.ly/3rchgod
- Bruno S, Coppola D, di Prisco G, Giordano D, Verde C (2019) Enzymes from Marine Polar Regions and Their Biotechnological Applications. Mar Drugs 17: 544. Link: https://bit.ly/3qiOnHX
- Garcia Moreno ML, Landgraf M (1998) Virulence factors and pathogenicity of Vibrio vulnificus strains isolated from seafood. J Appl Microbiol 84: 747-751. Link: https://bit.ly/3trOZwC
- Shields P, Cathcart L (2010) Oxydase Test Protocol. American Society for Microbiology. Link: https://bit.ly/3tpubGp
- Zaccone R, Caruso G, Calì C (2002) Heterotrophic bacteria in the northern Adriatic Sea: Seasonal changes and ectoenzyme profile. Mar Environ Res 54: 1-19. Link: https://bit.ly/3zOg8v3
- Kim SJ, Hoppe HG (1986) Microbial extracellular enzyme detection on agar plates by means of fluorogenic methylumbelliferyl-substrates. GERBAM-CNRS Actes des Colloques 3: 175–183. Link: https://bit.ly/3GjEuPS
- Hammer O, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software package for education and data analysis. Paleontol Electronica 4: 1-9. Link: https://bit.ly/3fcZiwo
- Oliver JD (1982) Taxonomic scheme for the identification of marine bacteria. Deep Sea Res 1 Oceanogr Res Pap 29: 795-798. Link: https://bit.ly/3I2LbGx
- Tropeano M, Coria S, Turjanski A, Cicero D, Bercovich A, Mac Cormack W, et al. (2012) Culturable heterotrophic bacteria from Potter Cove, Antarctica, and their hydrolytic enzymes production. Polar Res 31: 18507. Link: https://bit.ly/3tk4wyA
- Groudieva T, Kambourova M, Yusef H, Royter M, Grote R, et al. (2004) Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles 8: 475-488. Link: https://bit.ly/3GpO1or
- Fenice M, Selbmann L, Zucconi L, Onofri S (1997) Production of extracellular enzymes by Antarctic fungal strains. Polar Biol 17: 275-280. Link: https://bit.ly/3rdHIOv
- Dieser M, Greenwood M, Foreman CM (2010) Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct Antarct Alp Res 42: 396-405. Link: https://bit.ly/3Gghcdw
- Silva TRE, Silva LCF, de Queiroz AC, Alexandre Moreira MS, de Carvalho Fraga CA, de Menezes GCA, et al. (2021) Pigments from Antarctic bacteria and their biotechnological applications. Crit Rev Biotechnol 41: 809-826. Link: https://bit.ly/3fc2GYx
- Liu GY, Nizet V (2009) Color me bad: microbial pigments as virulence factors. Trends Microbiol 17: 406-413. Link: https://bit.ly/3ffFKYs

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
