Annals of Systems Biology
A Possible Path towards Prevention
Mihai Nadin*
Emeritus, The University of Texas at Dallas, USA
Cite this as
Nadin M. A Possible Path towards Prevention. Ann Syst Biol. 2024;7(1):054-056. Available from: 10.17352/asb.000024Copyright License
© 2024 Nadin M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Vaccination related to the COVID-19 pandemic turned out to be a global experiment. In the absence of clinical studies for assessing safety, dosage, immune response of subjects of different age groups, etc., vaccination as a means of reducing the number of victims practically replaced the final phase of the customary cycle of trials for FDA approval. Given this reality, methods for post-hoc evaluation need to be designed. This would serve as guidance for future preventive measures.
The experience accumulated during the pandemic (more or less 2019-2022) informs already a very vast array of research fields. Vaccinology is probably the most obvious, given the first large deployment of an mRNA-based vaccine. But epidemiology-mostly on account of testing means and methods-- comes very close, and so does medicine-where SARS-CoV-2 is no longer a matter of theoretic significance but one of life and death. Especially Long-COVID is in the process of becoming the dominant theme since a large number of people are affected, and medicine seems inadequate for addressing their specific ailments.
Against this background, we shall succinctly suggest a procedure for addressing various aspects of COVID-19, from a perspective that could benefit from the huge amount of data accumulated already. For instance, Australia detected its first cases of the highly transmissible XEC “recombinant” variant-which originated in Germany. It is a mix of two previous Omicron variants called KS 1.1 and KP 3.3. In the USA this variant affected over 10% of all COVID cases. Other countries are facing a similar situation. Updating vaccines, as new variants of the virus are detected and genetically described (a huge sequencing effort still in progress), is an example of an application of the procedure to be sketched in this short communication.
We started with the broader question of how variants are generated. The classic path for formulating a hypothesis is to discover causal inferences based on evidence [1]. Currently, evidence is derived from PCR (polymerase chain reaction) for identifying small amounts of virus material. It detects specific regions of the pathogen genome. Evidence is also derived from genetic analysis, in particular sequencing the whole viral genome (a technology still in progress). Indeed, Sanger sequencing [2], Capillary Electrophoresis (CE) [3,4], fragment analysis, Next generation sequencing (PGS), etc. are choices that make sense for different purposes. We shall not enter into the details informing the selection process. Suffice it to say that the “mechanics” of genetic processes, as it is called, returns only data regarding the chemistry of the process. The procedure we suggest is considering the timeline of interventions (vaccination, in particular) and juxtaposing it over the timeline of variant dynamics (when does a variant emerge) described by sequencing. It is possible, also, to search the data available for inferences from vaccines (repeat vaccines, boosters, etc.) to variants.
We analyzed COVID-19 vaccination data from the OurWorldInData website [5] and categorized the various vaccines into 4 main groups:
- mRNA vaccines: Pfizer/BioNTech, Moderna
- Adenovirus vector vaccines: Oxford/AstraZeneca, Johnson&Johnson, Sputnik V, etc.
- Inactivated virus vaccines: Sinopharm, Sinovac, Bharat Biotech, etc.
The data shows that 48 countries utilized adenovirus vector vaccines but did not administer any mRNA vaccines. India provides an example - they used vaccines like Covaxin and Oxford/AstraZeneca but did not deploy any mRNA options, likely due to supply limitations.
The question, which will continue to preoccupy us is: can we analyze the timeline (when and how many such vaccines were detected) and compare it to the timeline of the emergence of virus variants? This will also answer the question of whether some vaccines are vaccines or not [6].
To justify this procedure, the following deserves attention: replication is a characteristic of life. This was observed by those active in biology (e.g. Gregor Mendel) but also by those outside the field (John von Neumann). André Boivin [7] provided the grounding for what eventually became the mRNA method of vaccination: “the macromolecular deoxyribonucleic acids govern the building of macro-molecular ribonucleic acids, and, in turn, these control the production of cytoplasmic enzymes.” [8].
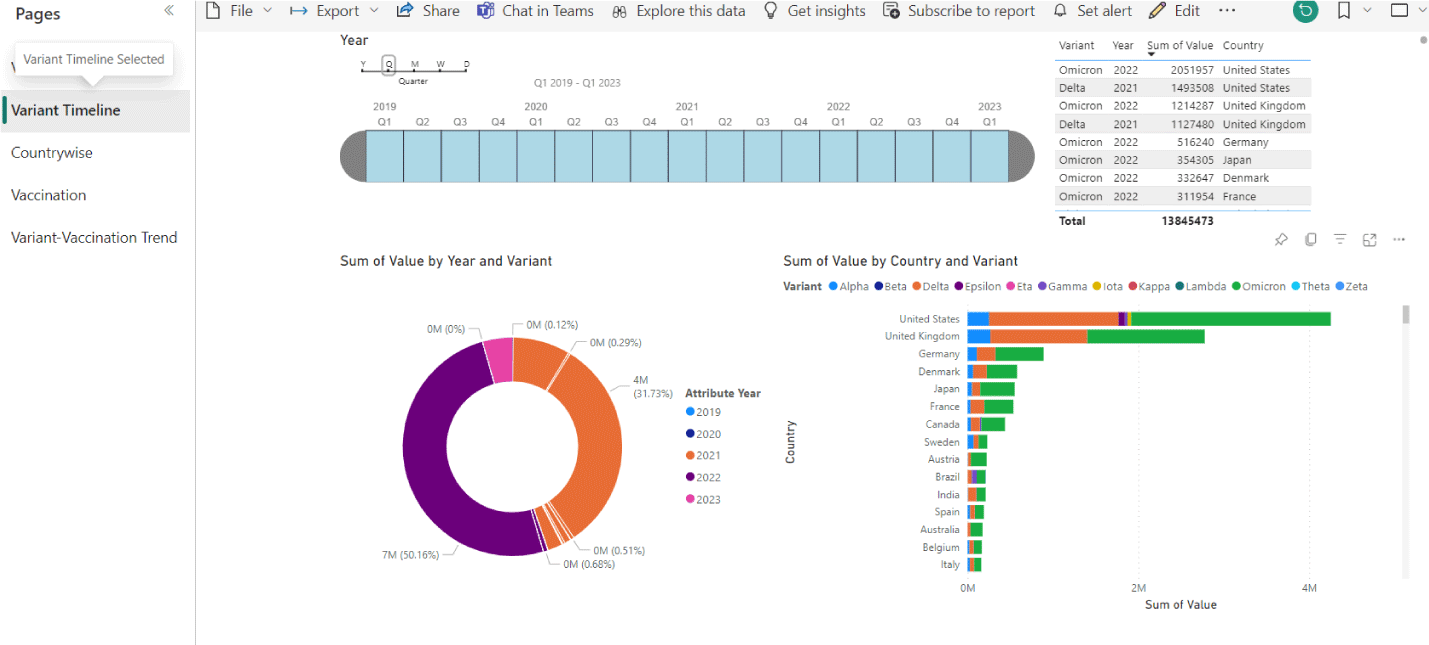
Ultimately, vaccines are substances (of natural or synthetic nature) used to trigger the production of antibodies and thus provide immunity against possible undesired conditions. But is it possible that in performing this function, they might change the originating of the disease? To find out if this is the case, we developed a procedure for searching the vast database database of performed vaccinations and that of the identified mutated virus (Figure 1). The procedure is used to analyze the relationship between variant emergence and vaccination rollout.
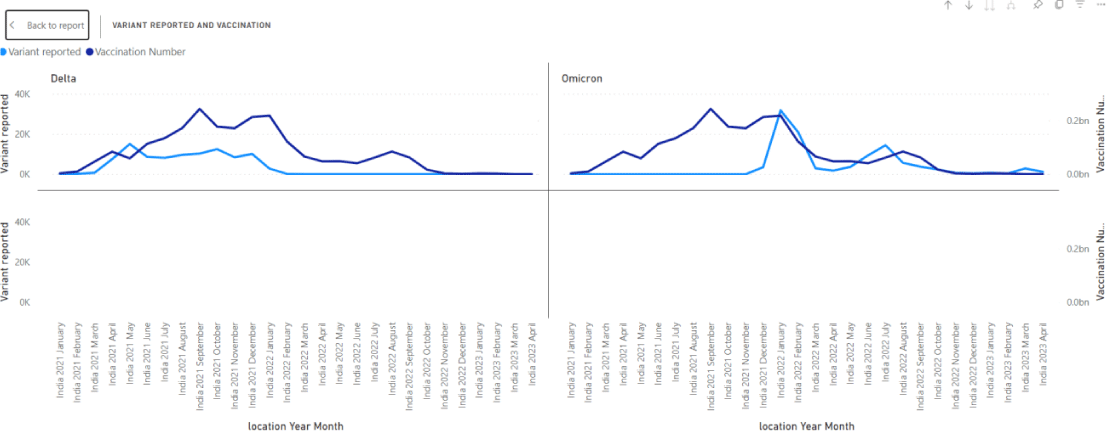
This work is in progress (Figure 2). So far, variant reports and vaccination give us a first indication that the effort is worth continuing. The hope is to eventually derive a predictive model (which would be useful in defining what kind of vaccination is appropriate).
Conclusion
The method presented in this short communication is only a beginning. The data made publicly available are too coarse to allow for predictive evaluations: Should subjects with compromised immune systems be treated the same way as healthy subjects are? Should children be vaccinated, given the low probability of their being infected and the relatively high incidents of myocarditis triggered by the mRNA vaccines? Of immediate practical interest would be to redefine the nature of data collected, the tools for analyzing data on the fly, and the possibility of providing context in the absence of which meaning is not accessible. Useful predictive models are the outcome of meaningful evaluation.
Acknowledging the contribution of Ayush Sharma and Shekhar Gaurav in the data processing effort.
- Stegenga J. Evidence of effectiveness. Stud Hist Philos Sci. 2022;91:288-295. Available from: https://doi.org/10.1016/j.shpsa.2022.01.001
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463-5467. Available from: https://doi.org/10.1073/pnas.74.12.5463
- Oda RP, Clark R, Katzmann JA, Landers JP. Capillary electrophoresis as a clinical tool for the analysis of protein in serum and other body fluids. Electrophoresis. 1997;18(10):1715-1723. Available from: https://doi.org/10.1002/elps.1150181004
- Sarker SD, Nahar L. An introduction to natural products isolation. In: Molecular Biology. Humana Press; 2012:864. Available from: https://doi.org/10.1007/978-1-61779-624-1_1
- Roser M. OWID homepage. Our World in Data. 2024 Mar 25. Available from: https://ourworldindata.org/
- Nadin M. Disrupt science. The Future Matters. Springer Cham; 2023. Available from: https://doi.org/10.1007/978-3-031-43957-5
- Boivin A, Vendrely R. On the possible role of the two nuclear acids in the living cell. Experientia. 1947;3:32-34. Available from: https://doi.org/10.1007/bf02155119
- Nadin M. Changing the definition does not make a vaccine more effective. Arch Pharm Pharmacol Res. 2023;3(3). Available from: https://irispublishers.com/appr/pdf/APPR.MS.ID.000562.pdf
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
