Annals of Marine Science
Case Report: First occurrence of Lymphocystis disease virus 3 (LCDV-Sa) in Wild Marine Fish in Tunisia
Nadia Cherif*, Fatma Amdouni, Kaouther Maatoug and Sami Zaafran
Cite this as
Cherif N, Amdouni F, Maatoug K, Zaafran S (2020) Case Report: First occurrence of Lymphocystis disease virus 3 (LCDV-Sa) in Wild Marine Fish in Tunisia. Ann Mar Sci 4(1): 024-029. DOI: 10.17352/ams.000022The results of the present study describe the molecular identification of Lymphocystis Disease Viral partial genome (LCDV-Sa) and histopathology in skin nodules and internal organs of Tunisian gilthead sea bream (Sparus aurata). The report supports as well as the existence of multiple reservoirs of LCDV within wild fish species caught near the cage facility; delivering a list of possible vector species susceptible to transmit the disease to farmed sea bream. Histology sections revealed irregular nucleus with basophilic cytoplasmic inclusions. In addition, to the best of our knowledge, molecular results added new viral reservoirs to the list of susceptible fish species described worlwide, including Sardinella aurita, Sardina pilchardus, Trachurus trachurus, Sarpa salpa, Diplodus vulgaris, Diplodus puntazzo, Liza aurata, Sparus aurata, Diplodus anularis, and Spicara maena. The virus detection was not correlated with neither the fish species nor the sampling temperature which varied between 15°C and 25°C. Partial sequence analysis of the MCP gene indicated that the newly identified LCDV strains were clustered within genotype VII and shared 96-100% of sequence identity with previously identified Tunisian LCDV sequences from farmed sea bream.
Introduction
Lymphocystis Disease (LD) is a rarely fatal, chronic and slowly developing disease, that affects over 150 different marine and fresh water fish species [1-8]. These include species that are of a particular importance for fish farming, such as Sparus aurata (gilthead sea bream). The typical sign of lymphocystis disease is the presence of small pearl-like nodules on the skin and fins of affected fish, that may occur singly or more generally grouped in raspberry-like clusters of tumorous appearance [9]. The external clinical signs of affected fish make them unmarketable [10] and more susceptible to other bacterial, viral or parasitic infections; increasing mortality rates and important economic loss. The etiological agent of LCD is the Lymphocystis Disease Virus (LCDV), a member of the genus Lymphocystivirus, family Iridoviridae. The Major Capsid Protein (MCP) gene represents an important molecular marker for the LCDV genotyping [11,12]. Data obtained on the basis of MCP sequences currently support the existence of nine genotypes in the genus Lymphocystis virus [13]. The genotype VII represents isolates obtained from sea bream and senegalese sole Solea senegalensis [14].
Tunisia imports more than 70% of the juvenile fish needed for marine cage farming. Currently, the aquatic bio-security and fish health management protocols used are minimal. Existing measures for disease prevention rely on the use of general prophylactic practices, such as good husbandry practices, reduced stocking density and enforcing the virological control of specimen to be introduced in the farming sites in order to detect carrier fish. This manuscript describes the detection of lymphocystis disease virus (LCDV) in gilthead sea bream, from both asymptomatic and diseased fish, collected at several Tunisian farms. It also describes the detection of LCDV in a number of asymptomatic specimens belonging to different wild fish species collected around farming facilities.
Materials and methods
During the routine auto-control for viral diseases by Tunisian fish farmers during 2015-2016, gilthead sea bream fish were randomly collected from four production facilities located at the Sahel region (Central-East coast), approximately 8 to10 kilometers far from each other. A total of 37 pooled samples were obtained from clinically ill and healthy Sparus aurata specimens, belonging to different growth stages were analyzed by molecular tools in order to detect LCDV. Samples included newly imported juveniles from different South European hatcheries. In addition, 96 wild fish specimens were caught around or inside S. auarta cages and processed in order to screen new viral reservoirs. Wild specimen species were identified based on Food and Agriculture Organization fish identification sheets [15].
Internal organs (liver, kidneys, and spleen), nervous tissues (eyes and brain), portions of skin and caudal fins, obtained individually from wild fish samples or pooled (5 specimen/pool) from farmed sea bream samples, suspended (1/10, w/v) in Leibovitz medium (L-15) supplemented with 2% FBS, 2% L-glutamine and 1% antibiotic solution (100 IU penicillin and100 mg streptomycin/mL), and subsequently homogenized (Table 1). For histological studies, the fins and skin nodules of infected sea bream fish were immediately preserved in 10% neutral-buffered formalin solution, sectioned transversally and longitudinally, exchanged to ethanol, dehydrated and embedded in paraffin blocks. Tissues were sectioned to 5 µm and stained with hematoxylin and eosin. The section samples were observed by light microscopy.
Total DNA was extracted from fish tissue homogenates using the QIAamp DNA Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. DNA quantity and purity (260/280 ratio) were estimated in a RNA/DNA Calculator (Nanodrop, thermo scientific). A DNA plasmid including a 609 bp region from the MCP gene corresponding with nucleotide positions 99 to 707 of the LCDV SA9 MCP gene (GenBank accession no. GU320728) was generously supplied by Dr. Dolores Castro (University of Malaga, Spain) and used in this study as a positive control. End point PCR assays were performed using the GoTaq Green Master Mix (Promega Madison, USA) in a total volume of 25 µl. The reaction contained 0.6 µM of each primer [10] and 50 ng of extracted DNA in addition to 10mM of dNTP, 50mM of Mgcl2, and 2,5µl of 10X enzyme buffer. Cycling parameters were: 1 min denaturation (95°C), 30 min annealing (50°C) and 1 min extension (72°C) for 35 cycles. The reaction was started by a denaturation step (2 min at 94 °C) and ended by a 5 min extension step at 72 °C. In every set of experiments, DNA from non-infected BF-2 cells was included as negative control. In a second time, a nested PCR reaction using an internal primer set LCDVm-F/LCDVm-R [16] was performed in order to improve the sensitivity of the diagnosis of the LCDV genome from asymptomatic fish specimens following the same amplification parameters as described in the first PCR. DNA products were analyzed on 1% agarose gels containing SYBR safe (Invitrogen), purified and sent to be sequenced. BLASTN analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was conducted with representative virus sequences exhibiting significant sequence. MCP gene nucleotide sequences of 39LCDV isolates available in GenBank were includedin the analyses.Pair wise comparison was performed using the blast2seqprogram (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi).Nucleotide partial sequences of the MCP gene were aligned via the MEGA6 [17] software using the Muscle method, and final adjustments were performed manually. The phylogenetic tree was constructed using the MEGA6 with UPGMA method and the final phylogenetic tree was drawn with the Coral DRAWX6 program. Because the sequences from GenBank were heterogeneous in length, the longer sequences were cut and adapted to the shortest. The reliability of the tree was inferred using the bootstrap method with 1000 replicates [18]. The partial nucleotide sequences of the LCDV strains screened in the present study were not deposited in Gen Bank because they are less than 200 pb.
Results
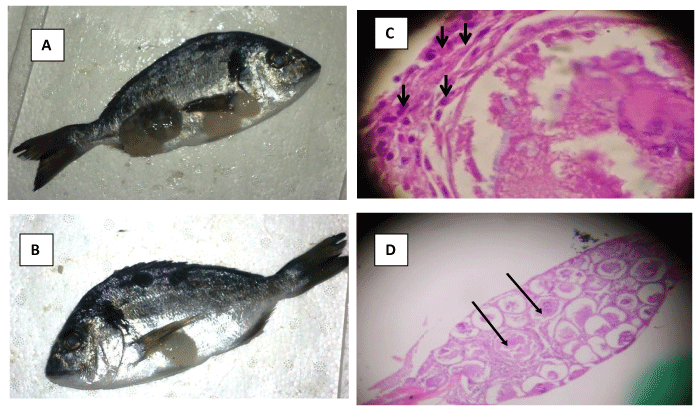
Histological results revealed that the lesions sampled from the skin and fins of infected S. aurata exhibited several characteristics that are known to be associated with LCD: ie, infected cells did not invade underlying muscular tissue, cells were severely hypertrophied, hyaline capsules clearly surround infected cells, and basophilic intra cytoplasmic inclusions are visible (Figures 1A-C). Histological sections stained with hematoxylin and eosin revealed irregular nucleus with margination of chromatin and basophilic intra-cytoplasmic inclusions (Figure 1D).
The identification of the wild fish specimen caught for the survey revealed 13 fish species belonging to five different families including the Sparidae, the Clupeidae, the Mugilidae, the Centracanthidae and the Carangidae, with no clinical signs.
The number of positive LCDV samples found within wild specimens at the studied locations is detailed in Table 2. Nested PCR results show that the virus was present at all the four geographical areas with a prevalence that varied between 34 to 44,7%. Positive wild fish species were as follows: Sardinella aurita, Sardina pilchardus, Trachurus trachurus, Sarpa salpa, Diplodus vulgaris, Diplodus puntazzo, Liza aurata, Sparus aurata, Diplodus anularis, and Spicara maena. The virus detection was not correlated with neither the fish species nor the sampling temperature which varied between 15°C and 25°C.
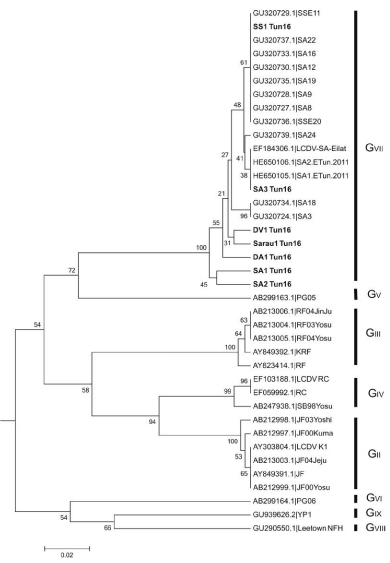
The comparison of three partial MCP gene sequences, obtained from farmed gilthead sea bream Sparus auarata (SA1-Tun16, SA2-Tun16 and SA3-Tun16), to the available reference sequences retrieved from Gene Bank (Table 2) showed nucleotide identities of 96–100% with the gilthead LCDV strains classified as genotype VII. Lower homology was obtained with sequences of other genotypes. Four sequences were obtained from wild fish specimens, as follows: Sarpa salpa (SS1-Tun16), Sparus aurata (Sarau1-Tun16), Diplodus vulgaris (DV1-Tun16) and Diplodus annularis (DA1-Tun16). The comparison of their partial MCP sequences exhibited 96-100% of sequence identity with LCDV genotype VII representatives as well. The phylogenetic tree based on MCP gene partial nucleotide sequences confirmed the classification of the Tunisian LCDV within the genotype VII, regardless the fish host and the geographic origin (Figure 2). The newly listed sequences were not deposited to a public access database as they were less than 200 nt.
Discussion
LCDV outbreaks are frequently observed in the Mediterranean gilthead sea bream aquaculture [19] even though, it is usually described as a self-limiting disease, there have been several reports on mortalities ranging up to 45 % in juvenile fish, which were related to secondary bacterial infections. Alternatively mortality may be linked to lymphocystis lesions which may severely impair fish respiration and/or feeding [20,21].
Very little proactive viral surveillance in North Africa there has been conducted, subsequently, few epidemiological data related to this virus in this region of the Mediteranean sea is available.
African Lymphocystis viral isolates have only been identified in Cichlids; including species of Tilapia in Lakes Victoria (Nyanza) (Oreochromis variabilis and Haplochromis spp.), in Lake George (H. elegans) and in Lake Kitangiri (Tilapia amphimelas and O. esculentus) in East Africa [22]. In Tunisia, LCDV has only been identified in farmed sea bream (S. aurata) specie [23].
The results of the present study support the existence of multiple reservoirs of LCDV at the farm facilities. To the best of our knowledge, this report describes the first identification of lymphocystis disease virus from wild Tunisian fish delivering a list of possible vector species susceptible to transmit the disease to farmed sea bream.
Genetic variations have been detected among LCDVs isolated from different hosts [10,24,25]. These affected species belong to evolutionarily advanced orders of teleosts fishes, including these families: Cichlidae, Osphronemidae, Centrarchidae, Gobiidae, Chaetodontidae, Pomacentridae, Sciaenidae, Serranidae and Pleuronectidae. As far as the authors are aware, LCDV has not been previously reported in less-advanced fish orders, such as Siluriformes, Cyprinids and Salmonids.
The Iridoviridae family has been determined to have highly conserved regions within the MCP gene [11] making it an ideal target for the identification of the virus. On the basis of LCDV Tunisian sequences that have been published to date, low genomic variability was observed among isolates from different fish farms spaced over 7 years. The survey indicates that the newly identified LCDV isolates were clustered within genotype VII and shared 96-100% of sequence identity with other Tunisian LCDV isolates previously identified in 2005 [13] and in 2011 [10]. Sequences were also closely related to strains isolated from other regions of south Europe [13]. This could be due to the active trade of fish among farms located in the Mediterranean Sea (Spain, Italy, France and Turkey), which make it difficult to draw any conclusion about the geographical distribution of the isolates. Full genome sequencing would allow a more thorough determination of the phylogenetic relationship between local and imported LCDV strains and could elucidate more about the viral origin of the Tunisian isolates.
The present work was funded by the Tunisian ministry of Agriculture (IRESA, 03/0013). We generously thank Dr. Dolores Castro for her precious help to initiate and revise this work by supplying the LCDV-positive control and unpublished PCR primers and protocols to our lab.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
- Anders K (1989) Lymphocystis disease of Fishes. viruses of lower vertebrates 141-160. Link: https://bit.ly/2Ty9HbK
- Marcogliese DJ, Fournier M, Lacroix A, Cyr DG (2001) Non-specific immune response associated with infections of lymphocystis disease virus in American plaice, Hippoglossoidesplatessoides (Fabricius). J Fish Dis 24: 121-124. Link: https://bit.ly/2LZMqeg
- Paperna I, Vilenkin M, de Matos AP (2001) Iridovirus infections in farm-reared tropical ornamental fish. Dis Aquat Organ 48: 17–25. Link: https://bit.ly/2TBoB0E
- Bunkley-Williams L, Williams EH, Phelps RP (2002) Does lymphocystis occur in pacora, Plagioscionsurinamensis (Sciaenidae), from Colombia? Acta Tropica 82: 7–9. Link: https://bit.ly/2ZxI06t
- Sheng XZ, Zhan WB, Wang Y (2007) Whitespotter puffer Arothronhispidus, a new host for lymphocystis in Qingdao Aquarium of China. Diseases of Aquatic Organisms 75: 23–28. Link: https://bit.ly/2A2z3HI
- Hossain M, Song JY, Kitamura SI, Jung SJ, Oh MJ (2008) Phylogenetic analysis of lymphocystis disease virusfrom tropical ornamental fish species based on a major capsidprotein gene. J Fish Dis 31: 473–479. Link: https://bit.ly/3c45QJI
- Xu L, Feng J, Huang Y (2014) Identification of lymphocystis disease virus from paradise fish Macropodusopercularis (LCDVPF). Arch Virol 159: 2445–2449. Link: https://bit.ly/2LSgVmC
- Huang X, Huang Y, Xu L, Wei S, Ouyang Z, Feng J (2015) Identification and characterization of a novel lymphocystis disease virus isolate from cultured grouper in China. J Fish Dis 38: 379–387. Link: https://bit.ly/36sI9tj
- Smail DA, Munro ALS (2001) The virology of teleosts. In: Roberts RJ, editor. Fish pathology. 3rd ed. Edinburgh: W.B. Saunders;p. 169–253.
- Kitamura SI, Jung SJ, Kim WS, Nishizawa T, Yoshimizu M, OhM J (2006) A new genotype of Lymphocystisvirus, LCDV-RF, from lymphocystis diseased rockfish. Arch Virol 151: 607-615. Link: https://bit.ly/2XsC500
- Tidona CA, Schnitzler P, Kehm R, Darai G (1998) Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16: 59- 66. Link: https://bit.ly/3ghoNff
- Williams T, Barbosa-Solomieu V, Chinchar VG (2005) A decade of advances in iridovirus research. Virus Res 65: 173- 248. Link: https://bit.ly/36rpHS1
- Palmer LJ, Hogan NS, Van den Heuvel MR (2012) Phylogenetic analysis and molecular methods for the detection of lymphocystis disease virus from yellow perch, Percaflavescens (Mitchell). J Fish Dis 35: 661-670. Link: https://bit.ly/2A2u4qD
- Cano I, Valverde EJ, Lopez-Jimena B, Alonso MC, Garcia-Rosado E, et al. (2010) A new genotype of Lymphocystivirus isolated from cultured gilthead seabream, Sparus aurata L., and Senegalese sole, Solea senegalensis (Kaup). J Fish Dis 33: 695-700. Link: https://bit.ly/2yvumWx
- Fisher W Bauchot ML, Schneider M (1987) Fiches F.A.O. Identification des espèces pour les besoins de la pêche "Révision 1" Méditerranée et Mer noire. Zone de pêche 37. Volume I. Invertébrés marins. Rome, FAO 2: 761 - 1530.
- Valverde EJ, Cano I, Labella A, Borrego JJ, Castro D (2016) Application of a new real-time polymerase chain reaction assay for surveillance studies of lymphocystis disease virus in farmed gilthead seabream. BMC Vet Res 12: 71. Link: https://bit.ly/3gl8aiU
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. Link: https://bit.ly/3d1WBei
- Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791. Link: https://bit.ly/2XudqIa
- Borrego JJ, Castro D, Balebona MC, Garcia-Rosado E, Lopez-Cortes L (2001) Patologı´as que Afectan al Cultivo de la Dorada (Sparusaurata, L.) en la ComunidadAuto´nomaAndaluza. Consejerı´a de Agricultura y Pesca, Junta de Andalucı´a, Sevilla.
- Colorni A, Padros F (2011) Diseases and health management. Edited byPavlidis MA andMylonas CC.Sparidae: Biology and Aquaculture of Gilthead Sea Bream and other Species, Wiley-Blackwell, Oxford 321-357.
- Dezfuli BS, Lui A, Giari L, CastaldelliMulero V, Noga EJ (2012) Infiltration and activation of acidophilic granulocytes in skin lesions of gilthead seabream, Sparus aurata, naturally infected with lymphocystis disease virus. Dev Comp Immunol 36: 174-182. Link: https://bit.ly/2LUAM4m
- Abowei JFN, Briyai OF, Bassey SE (2011) A Review of Some Viral, Neoplastic, Environmental and NutritionalDiseases of African FishBritish Journal of Pharmacology and Toxicology 2: 227-235.
- Haddad-Boubaker S, Bouzgarou N, FakhfakhE, Khayech M, Ben Mohamed S, et al. (2013) Detection and GeneticCharacterization of Lymphocystis Disease Virus (LCDV) Isolated during Disease Outbreaks in Cultured Gilt-head Sea Bream Sparus aurata in Tunisia. Fish Pathology 48: 101-104. Link: https://bit.ly/2ZxYPyj
- Tidona CA, Darai G (1997) The complete DNA sequence of lymphocystis disease virus. Virology 230: 207-216. Link: https://bit.ly/3ehkrDb
- Zhang QY, Feng X, Jian X, Li ZQ, Gui JF (2004) Complete Genome Sequence of Lymphocystis Disease Virus Isolated from China. J Virol 5: 6982-6994. Link: https://bit.ly/36wzgPI

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
